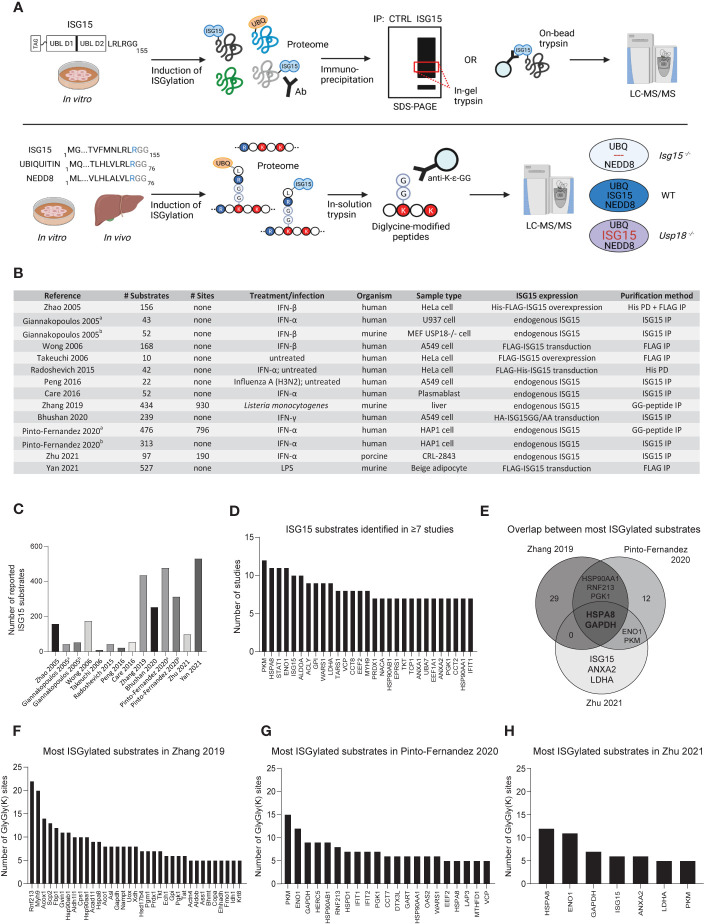
Figure 2.
MS-based discovery of ISG15 substrates. (A) Commonly used workflows to identify ISG15 substrates by mass spectrometry. Upper panel: many studies rely on pulldown of ISGylated proteins from induced cells using antibodies or resins against endogenous or overexpressed (tagged) ISG15. Pulled down proteins are separated by SDS-PAGE to trypsinize specific gel bands. Alternatively, proteins are digested directly on-bead. In both cases, the released peptides are analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Non-specific binders can be controlled by parallel analysis of cells expressing no ISG15 or non-conjugatable ISG15 (e.g. ISG15AA) or by using an antibody isotype control. Lower panel: to identify exact sites of ISG15 modification, recent studies applied diglycine (GG) peptide enrichment from induced cells or tissues. Here, trypsin digestion of extracted proteins generates GG-modified peptides derived from conjugated ISG15, ubiquitin or NEDD8. These GG-modified peptides can be captured with specific anti-K-ϵ-GG antibodies prior to their identification by LC-MS/MS. To distinguish ISG15 modification sites from ubiquitin and NEDD8 sites, genetic controls are included such as Isg15-/- mice or Usp18-/- cells. Consequently, comparison with a wild-type condition allows to pinpoint bona fide ISG15 sites based on their absence in Isg15-/- or increased abundance in Usp18-/- conditions. Figure created with BioRender.com. (B) Overview of MS-based proteomics studies reporting ISG15 substrates, listing relevant information on the number of identified substrates and the experimental design. (C) Bar chart showing the increase of identified ISG15 substrates in the different studies over time. (D) Bar chart showing 29 ISG15 substrates that were reported in ≥7 studies. (E) Venn diagram showing the overlap between the most modified ISG15 substrates listed in panel (F–H). (F–H) Bar charts showing the most modified ISG15 substrates (substrates with ≥5 sites) in the three GG-peptidomics screens.