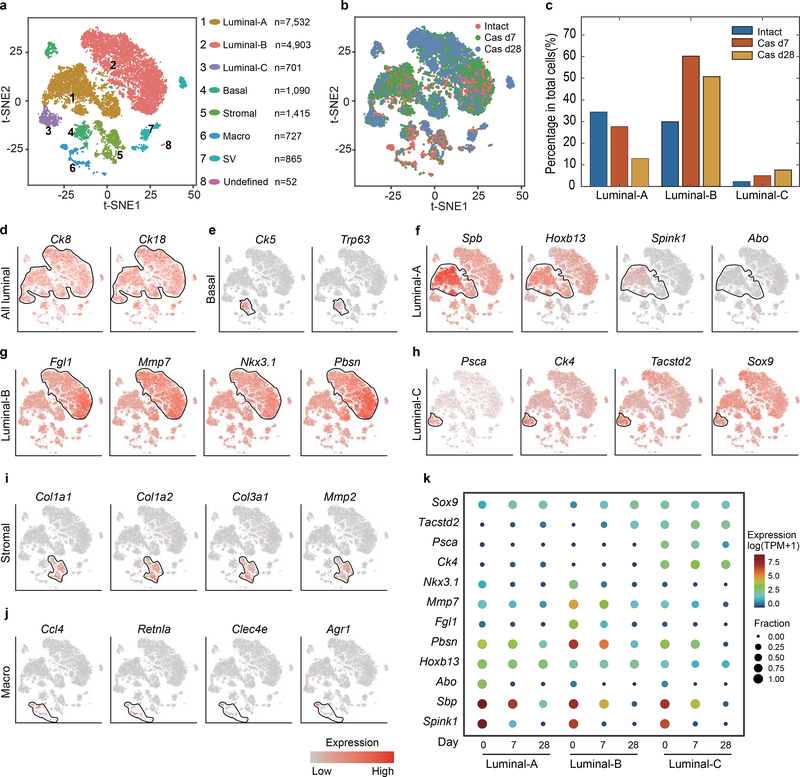
Extended Data Fig. 6 |. Single-cell transcriptomic survey for prostate cells of intact and regressed mice (castrated for 7 days and castrated for 28 days).
a, Visualization of clustering of 17,305 single cells (points; n = 6 mice), based on the expression of known marker genes by t-SNE (left panel). Cell numbers and percentages of assigned cell types are summarized in the right panel. Luminal-A, type A luminal cell; Luminal-B, type B luminal cell; Luminal-C, type C luminal cell; Basal, basal cell; Stromal, stromal cell; Macro, macrophage; SV, seminal vesicle epithelial cell. b, T-SNE map shows the cell distribution from intact mice (day 0; n = 8,545 cells), regressed mouse (day 7; n = 5,734 cells) and (day 28; n = 3,006 cells). c, Bar graph shows the percentage of Luminal-A, Luminal-B and Luminal-C cells in total cells of intact mice (day 0), regressed mouse (day 7) and (day 28). d-j, T-SNE maps show the expression levels of marker genes across 8 clusters. Black circles indicate all luminal cluster (n = 13,136 cells) (d), basal cluster (n = 1,090 cells) (e), the Luminal-A cluster (n = 7,532 cells) (f), Luminal-B cluster (n = 4,903 cells) (g), Luminal-C cluster (n = 701 cells) (h), stromal cluster (n = 1,415 cells) (i), macrophage cluster (n = 727 cells) (j). t-SNE map shows cells that are colored by the log-scale normalized read count of marker genes. k, Mean Expression in target cells (log2[TP10K], color) and fraction of expressing cells (dot size) of key luminal markers of Luminal-A, Luminal-B and Luminal-C (rows) in cells from each luminal sub-clusters of intact mice (day 0), regressed mouse (day 7) and (day 28) (columns).