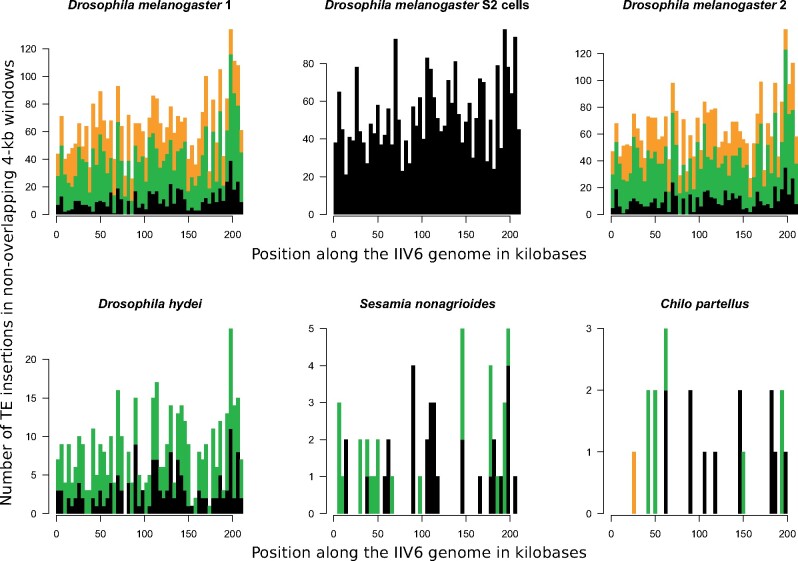
Fig. 4.
Stacked histograms showing the number of different insertions of TEs in 4-kb windows along the genome of the iridovirus IIV6 in a parental viral population (purified from Drosophila melanogaster S2 cells) and five IIV6 daughter populations purified from other hosts. The top central diagram illustrates TE insertions in the parental IIV6 purified from D. melanogaster S2 cells. The other five diagrams illustrate TE insertions in the daughter IIV6 populations purified from whole flies or moths (fig. 1). Black bars correspond to TE insertions shared by the parental IIV6 population and at least one daughter IIV6 population. These shared insertions correspond to the same TE at the exact same position in the parental IIV6 population purified from D. melanogaster S2 cells and other IIV6 populations purified from other hosts. Green bars correspond to TE insertions not shared with the parental IIV6 population. TEs involved in these unshared insertions were present in the parental IIV6 population but not at the position they were found in the daughter populations. These unshared insertions may be due to de novo virus-to-virus transposition during infection of Drosophila whole flies or moth larvae. Alternatively, they may correspond to TE insertions that were present in the parental population but were not detected by sequencing. Orange bars correspond to TEs that are absent from the parental IIV6 population. These are likely to result from de novo transposition from the host genome. Numbers of insertions are given in supplementary table S1, Supplementary Material online, for each TE.