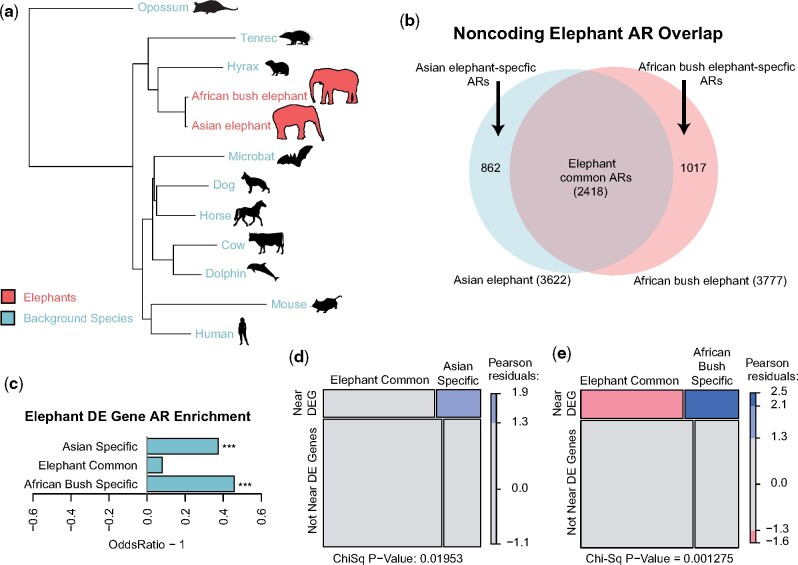
Fig. 1.
Accelerated regions in elephant genomes. (a) Using a whole genome alignment of 12 mammals we defined genomic regions that were accelerated (ARs) in two elephant species (red), yet conserved in the set of background species (blue). Branch lengths are given in terms of mean substitutions per site at 4-fold degenerate sites (neutral model). (b) Among the ARs detected in elephants, we found ARs common to both elephant species as well as ARs specific to either Asian or African bush elephants. (c) Differentially expressed (DE) genes were much more likely to be found in Asian elephant-specific (Fisher’s Exact Test, P = 2.05e−4) and African elephant-specific (P = 8.30e−7) ARs than in common ARs. (d, e) Species-specific ARs disproportionately overlap DE gene regulatory regions relative to the common ARs (χ2 test, P = 0.019 and P = 0.001, respectively).