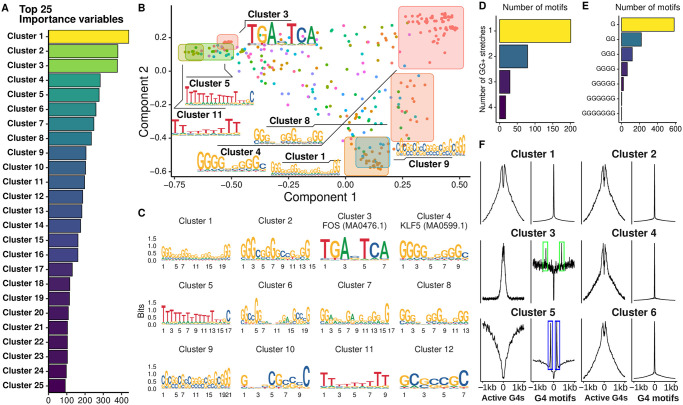
Fig 4. DNA motifs identified by DeepG4.
A) Variable importances of DeepG4 cluster motifs, as estimated by random forests. Clustering of DeepG4 kernel motifs was done by RSAT matrix-clustering program to obtain cluster motifs. B) Multidimensional scaling (MDS) of DeepG4 motifs. As an input, matrix-clustering correlation matrix between kernel motifs was used. C) Logos of cluster motifs with highest variable importances. D) Number of kernel motifs containing one or more GG+ stretches. A GG+ stretch is defined as a stretch of 2 or more Gs in the motif consensus sequence. E) Number of kernel motifs containing G stretches depending on stretch length. F) Average profiles measuring the enrichment of cluster motifs centered around active G4 regions or canonical G4 motifs.