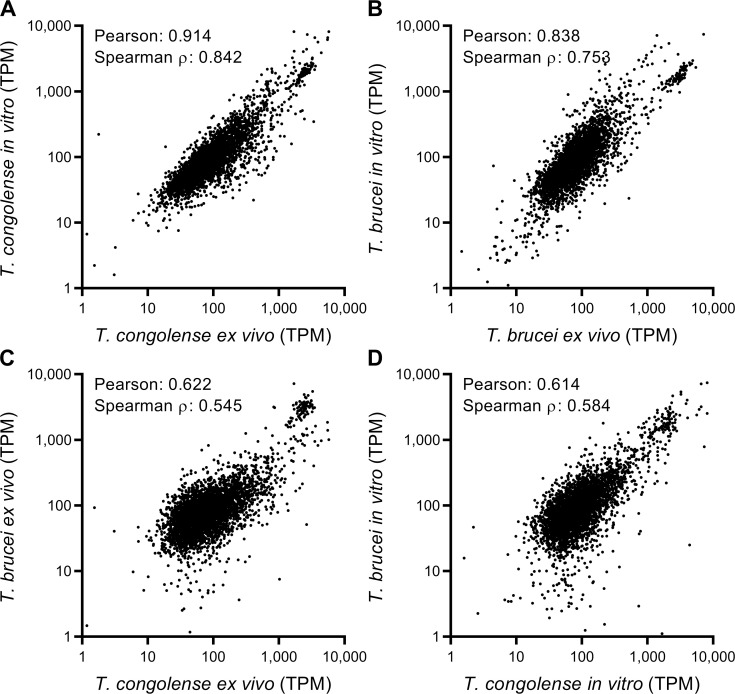
Fig 1. Overview of comparative transcriptomics analysis of T. brucei and T. congolense, isolated from ex vivo and in vitro conditions.
RNAseq data from T. congolense (IL3000) and T. brucei (STIB 247) in both in vitro and ex vivo (from mouse infections) conditions were aligned to the species’ respective reference genomes and read counts were normalised by the transcripts per million (TPM) method. To directly compare the species, a pseudogenome was generated using the Orthofinder tool [57]. TPM values from the 4 sample groups were plotted against each other to analyse correlation between conditions (A and B) and between species in the same conditions (C and D). Correlation was assessed using both Pearson correlation (Pearson’s r) and Spearman’s rank correlation coefficients.