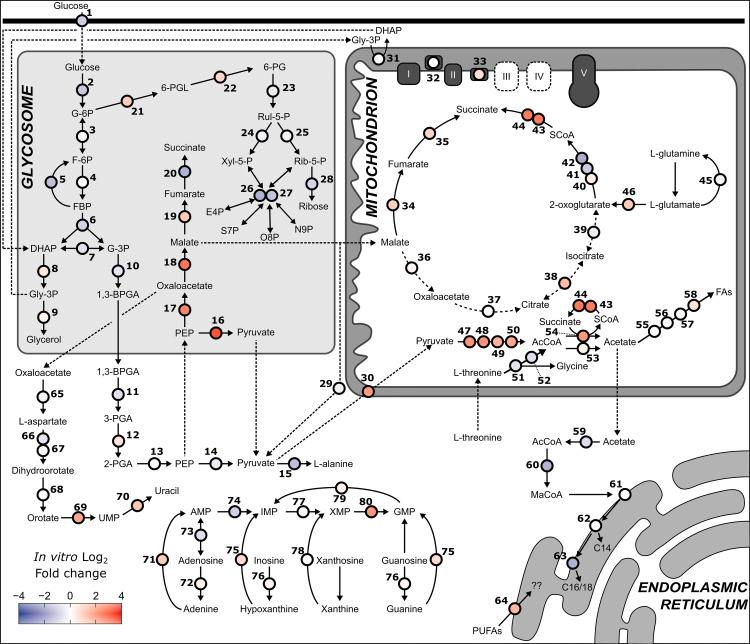
Fig 8. Summary of T. congolense and T. brucei in vitro transcriptome.
Log2 fold change (T. congolense/T.brucei) was calculated for each gene. Dashed lines represent transport processes. Genes: 1, hexose transporters, TbTc_0095; 2, hexokinase, TbTc_0341; 3, glucose-6-phosphate isomerase, TbTc_1840; 4, phosphofructokinase, TbTc_1399; 5, fructose-1,6-bisphosphatase, TbTc_1967; 6, aldolase, TbTc_0358; 7, triosephosphate isomerase, TbTc_1075; 8, glycerol-3-phosphate dehydrogenase, TbTc_2722; 9, glycerol kinase, TbTc_0392; 10, glyceraldehyde 3-phosphate dehydrogenase, TbTc_0377; 11, phosphoglycerate kinase, TbTc_0240; 12, phosphoglycerate mutase, TbTc_5039; 13, enolase, TbTc_0465; 14, pyruvate kinase 1, TbTc_0372; 15, alanine aminotransferase, TbTc_0675; 16, pyruvate phosphate dikinase, TbTc_1304; 17, Phosphoenolpyruvate carboxykinase, TbTc_0348; 18, glycosomal malate dehydrogenase, TbTc_0642; 19, glycosomal fumarate hydratase, TbTc_0242; 20, glycosomal NADH-dependent fumarate reductase, TbTc_0140; 21, glucose-6-phosphate dehydrogenase, TbTc_0931; 22, 6-phosphogluconolactonase, TbTc_4165; 23, 6-phosphogluconate dehydrogenase, TbTc_2025; 24, ribulose-5-phosphate epimerase, TbTc_4356; 25, ribose 5-phosphate isomerase, TbTc_3090; 26, transketolase, TbTc_1701; 27, transaldolase, TbTc_1823; 28, ribokinase, TbTc_5212; 29, malic enzyme, TbTc_0296; 30, Mitochondrial pyruvate carrier 2, TbTc_2668; 31, FAD-dependent glycerol-3-phosphate dehydrogenase, TbTc_2282; 32, NADH dehydrogenase (NDH2), TbTc_5033; 33, Alternative oxidase, TbTc_6589; 34, mitochondrial fumarate hydratase, TbTc_0243; 35, mitochondrial NADH-dependent fumarate reductase, TbTc_0141; 36, mitochondrial malate dehydrogenase, TbTc_0256; 37, citrate synthase, TbTc_0486; 38, aconitase, TbTc_5765; 39, isocitrate dehydrogenase, TbTc_0510; 40, 2-oxoglutarate dehydrogenase E1 component, TbTc_2864; 41, 2-oxoglutarate dehydrogenase E1 component, TbTc_3111; 42, 2-oxoglutarate dehydrogenase E2 component, TbTc_3057; 43, succinyl-CoA synthetase α, TbTc_0813; 44, succinyl-CoA ligase β, TbTc_3392; 45, glutamine synthetase, TbTc_2226; 46, glutamate dehydrogenase, TbTc_0872; 47, pyruvate dehydrogenase E1 α subunit, TbTc_4169; 48, pyruvate dehydrogenase E1 β subunit, TbTc_5437; 49, dihydrolipoamide acetyltransferase, TbTc_1015; 50, pyruvate dehydrogenase complex E3, TbTc_4765; 51, L-threonine 3-dehydrogenase, TbTc_5991; 52, 2-amino-3-ketobutyrate coenzyme A ligase, TbTc_6236; 53, Acetyl-CoA hydrolase (ACH), TbTc_5515; 54, Succinyl-CoA:3-ketoacid coenzyme A transferase (ASCT), TbTc_0236; 55, Acyl carrier protein, TbTc_5262; 56, beta-ketoacyl-ACP synthase, TbTc_3372; 57, beta-ketoacyl-ACP reductase, TbTc_1241; 58, Trans-2-enoyl-ACP reductase 1, TbTc_5269; 59, acetyl-CoA synthetase, TbTc_0318; 60, acetyl-CoA carboxylase, TbTc_0754; 61, Fatty acid elongase (ELO1), TbTc_0159; 62, Fatty acid elongase (ELO2), TbTc_1882; 63, Fatty acid elongase (ELO3), TbTc_0235; 64, elongation of very long chain fatty acids protein (ELO4), TbTc_0737; 65, aspartate aminotransferase, TbTc_0799; 66, aspartate carbamoyltransferase, TbTc_1630; 67, dihydroorotase, TbTc_3801; 68, dihydroorotate dehydrogenase, TbTc_0620; 69, orotidine-5-phosphate decarboxylase/orotate phosphoribosyltransferase, TbTc_0735; 70, uracil phosphoribosyltransferase, TbTc_4220; 71, Adenine phosphoribosyltransferase (APRT-2), TbTc_3522; 72, inosine-adenosine-guanosine-nucleoside hydrolase, TbTc_4998; 73, adenosine kinase, TbTc_1024; 74, AMP deaminase, TbTc_5808; 75, hypoxanthine-guanine phosphoribosyltransferase (HGPRT), TbTc_0726; 76, inosine-guanine nucleoside hydrolase, TbTc_0808; 77, inosine-5’-monophosphate dehydrogenase, TbTc_1648; 78, Hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), TbTc_3696; 79, GMP reductase, TbTc_4627; 80, GMP synthase, TbTc_1452. Abbreviations: PUFA, polyunsaturated fatty acid.