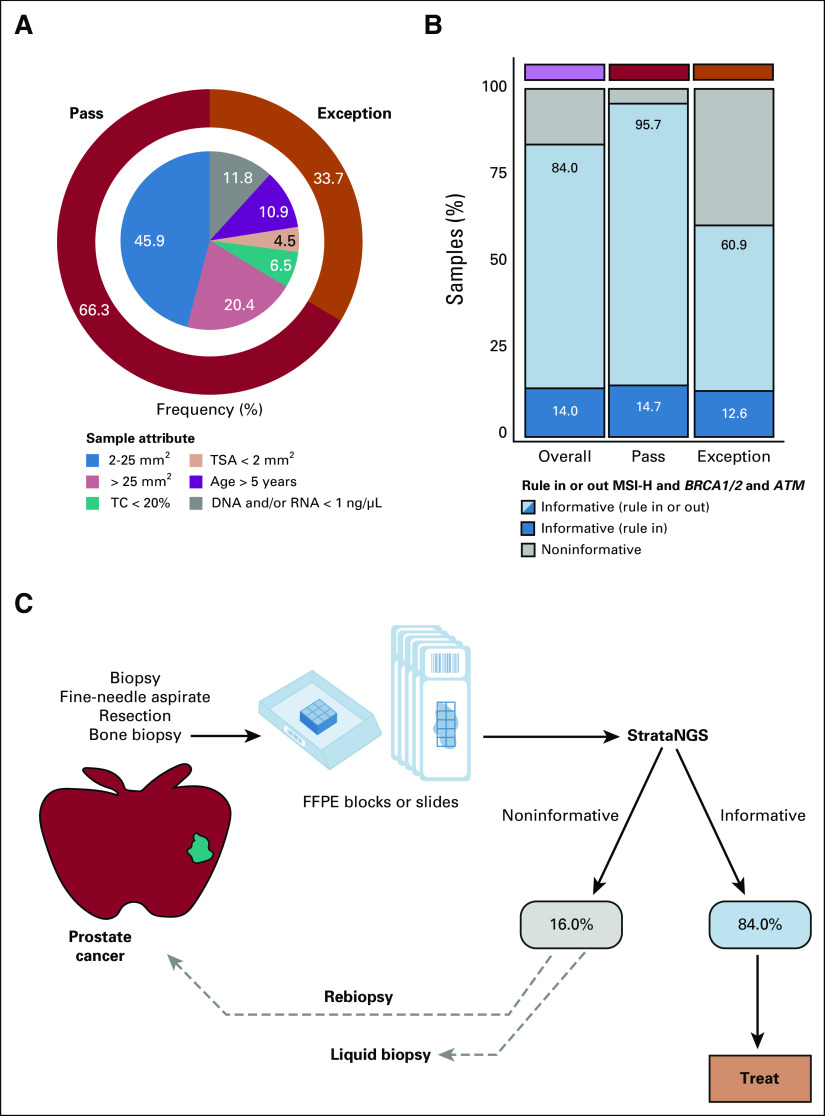
FIG 4.
(A) Donut plot characterizing the composition of consecutively tested, sample input characteristic-evaluable prostate cancer samples (n = 1,344) from the overall PCR-CGP test cohort. The outer ring indicates the percentage of samples meeting (pass: dark red or not meeting (exception: orange) PCR-CGP input requirements. In the inner pie chart, samples passing all input requirements are stratified by TSA; exception samples are stratified by indicated sample attribute (TC < 20%; TSA < 2 mm2; age > 5 years: specimen collected > 5 years before PCR-CGP; and DNA and/or RNA concentration < 1 ng/µL). (B) The proportion of tested samples (overall and by sample input requirement category) for which an informative result (able to rule in or out actionable alterations) was reported. To be considered informative (total of light and dark blue), the test must have reported (1) either MSI-H or a deleterious mutation and/or copy number deep deletion in MSH2/6, BRCA1/2, or ATM (dark blue) or (2) tested definitively negative for these biomarkers by meeting all sequencing QC metrics and having TC ≥ 20%, the PCR-CGP test's overall limit of detection (light blue). The percent of total informative and informative rule in are indicated. (C) Potential real-world prostate cancer testing paradigm on the basis of sample characteristics and PCR-CGP performance characteristics observed in this cohort. Patients with noninformative test results in this cohort were not followed to determine whether rebiopsy or liquid biopsy testing was pursued. CGP, comprehensive genomic profiling; FFPE, formalin-fixed paraffin-embedded; MSI-H, microsatellite instability–high; PCR-CGP, multiplex polymerase chain reaction–based comprehensive genomic profiling; QC, quality control; TC, tumor content; TSA, tumor surface area.