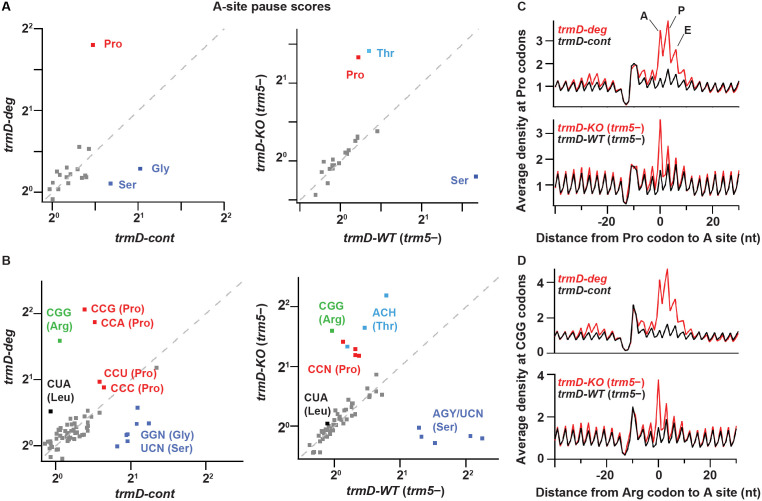
Figure 2. TrmD depletion leads to ribosome pausing at CCN (Pro) and CGG (Arg) codons.
(A) Pause scores for codons positioned in the ribosomal A site. The codons are grouped by amino acid. Left panel: data from biological replicates of trmD-deg and trmD-cont strains upon ClpXP induction. Each biological replicate is an independent culture of cells. Right panel: data from trmD-KO (trm5–) and trmD-WT (trm5–) strains upon turning off trm5. (B) The same as above but showing the 61 sense codons individually. (C) Plots of average ribosome density aligned at Pro codons for trmD-deg strain (top) and trmD-KO (trm5–) strain (bottom) in red with their respective control strains in black. The peaks corresponding to the ribosomal A, P, and E sites are labeled. (D) Plots of average ribosome density aligned at CGG codons as above. Note that trmD-deg samples were obtained with the traditional ribosome profiling lysis buffer, whereas trmD-KO (trm5–) samples were obtained with a high Mg2+ lysis buffer that more effectively arrests translation after cell lysis, revealing these pauses at higher resolution.