Summary
Rice sheath blight (RSB) is an economically significant disease affecting rice yield worldwide. Genetic resistance to RSB is associated with multiple minor genes, with each providing a minor phenotypic effect, but the underlying dominant resistance genes remain unknown. A genome‐wide association study (GWAS) of 259 diverse rice varieties, with genotypes based on a single nucleotide polymorphism (SNP) and haplotype, was conducted to assess their sheath blight reactions at three developmental stages (seedlings, tillering and booting). A total of 653 genes were correlated with sheath blight resistance, of which the disease resistance protein RPM1 (OsRSR1) and protein kinase domain‐containing protein (OsRLCK5) were validated by overexpression and knockdown assays. We further found that the coiled‐coil (CC) domain of OsRSR1 (OsRSR1‐CC) and full‐length OsRLCK5 interacted with serine hydroxymethyltransferase 1 (OsSHM1) and glutaredoxin (OsGRX20), respectively. It was found that OsSHM1, which has a role in the reactive oxygen species (ROS) burst, and OsGRX20 enhanced the antioxidation ability of plants. A regulation model of the new RSB resistance though the glutathione (GSH)‐ascorbic acid (AsA) antioxidant system was therefore revealed. These results enhance our understanding of RSB resistance mechanisms and provide better gene resources for the breeding of disease resistance in rice.
Keywords: rice sheath blight, Rhizoctonia solani AG1‐IA, genome‐wide association study, resistance genes, glutathione‐ascorbic acid antioxidant system
Introduction
Rice (Oryza sativa L.) is one of the world’s most important cereal crops, providing a key part of the daily dietary intake for approximately 50% of the global population (Chen et al., 2014). Rice sheath blight (RSB), a major disease of rice, is caused by the soil‐borne necrotrophic fungus Rhizoctonia solani Kühn of the anastomosis group AG1‐IA, and results in major losses in the yield of rice under favourable conditions (Gautam et al., 2003; Lee and Rush, 1983). RSB resistance is a very complex quantitative trait involving dynamic and diverse responses that are controlled by multiple genes (Zuo et al., 2000). The mechanisms of rice resistance to R. solani AG1‐IA remain largely unknown, and no dominant RSB resistance genes or loci have been discovered.
With the advent of high‐throughput genotyping technologies, such as resequencing and microarrays, a genome‐wide association study (GWAS) has become a powerful tool for the identification of genes associated with disease resistance in plants. For example, GWAS has been used to demonstrate the role of ZmFBL41, which encodes an F‐box protein, in the resistance to banded leaf and sheath blight in maize (Li et al., 2019). In rice, 27 rice blast resistance loci (Li et al., 2019) and 19 loci associated with resistance to bacterial blight have been reported (Dilla‐Ermita et al., 2017; Zhang et al., 2017a). Using 299 rice varieties, a recent study identified two genomic regions, qSB‐3 and qSB‐6, that are associated with R. solani resistance (Chen et al., 2019). Although these studies did identify candidate genes, in most cases they did not verify new resistance genes using transgenic methods.
The plant recognized pathogen effectors, such as nucleotide‐binding site leucine‐rich repeat (NLR) or receptor‐like cytoplasmic kinases (RLCK) proteins, have critical roles in the regulation of the immune response (Jia et al., 2000; Shao et al., 2003). In Arabidopsis thaliana, the RLCK protein PBS1 regulates resistance against Pseudomonas syringae and recognition by the P. syringae AvrPphB effector protease (Shao et al., 2003). Plant genomes encode several hundred NLR proteins involved in defence responses (Marone et al., 2013), some of which occur in clusters at specific loci following gene duplication and amplification events (Liu et al., 2014; Zhou et al., 2019). Furthermore, based on physical or genetic mapping it is apparent that many NLR genes co‐localize with resistance loci (McHale et al., 2006). Previous studies have shown that genetically linked NLR genes act together to recognize pathogen avirulence effectors, such as RPS4/RPS1, RGA4/RGA5 and Pikp‐1/Pikp‐2 (Cesari et al., 2013; Yuan et al., 2011; Zhang and Gassmann, 2003). In these pairs, one gene is a sensor that perceives pathogen effectors, while the other is a helper required for activating immune signalling (Baggs et al., 2017; Cesari et al., 2013; Wu et al., 2015). Although the role of NLR and RLCK proteins in plant immunity has been well studied, there have been no reports indicating the role of NLR or RLCK proteins in RSB resistance.
Plant defence responses against pathogen infection usually involve the activation of defence‐related hormone pathways, such as jasmonate acid (JA), salicylic acid (SA) and ethylene (ET); up‐regulation of pathogenesis‐related (PR) genes; and the production of reactive oxygen species (ROS) (van Loon et al., 2006). While the interaction mechanism of rice and R. solani AG1‐IA remains unclear (Pinson et al., 2005; Zou et al., 2000; Zuo et al., 2007), the regulation of JA‐dependent defence signalling and ROS has an important role in the resistance to necrotrophic fungi (Oreiro et al., 2019; Zhang et al., 2017b). It has been reported that ROS are involved in triggering plant basal defence responses to biotrophic and hemi‐biotrophic pathogens, as well as necrotrophs. For example, the NLR gene Pit imparts resistance against the rice blast fungus through mediated ROS production (Kawano et al., 2010).
In this study, 259 rice lines were evaluated for the response to RSB at three developmental stages. We used single nucleotide polymorphism (SNP)‐based (SNP‐GWAS) and haplotype‐based GWAS (Hap‐GWAS) to identify loci associated with RSB resistance via high‐density SNP genotyping. Core candidate genes were obtained through the weighted co‐expression network analysis (WGCNA) of candidate genes identified by SNP‐GWAS, Hap‐GWAS and a transcriptome analysis of resistant and susceptible varieties. Furthermore, we demonstrated how the disease resistance protein RPM1 encoding gene OsRSR1 and the protein kinase gene OsRLCK5 are involved in sheath blight resistance though the glutathione (GSH)‐ascorbic acid (AsA) antioxidant system. These findings set a stage to uncover the mechanism of plant innate immunity leading to RSB resistance.
Results
Genomic variation and population structure
To map the RSB resistance genes, we sequenced 259 rice varieties at an average coverage of 12.16 × using the Illumina HiSeq platform (Figure S1 and Table S1), generating 1.33 Tb of raw reads (Table S2). The sequence reads were mapped on to the Nipponbare reference genome, and 2,888,332 high‐confidence SNPs (missing data < 20%, minor allele frequency [MAF]>1%) were detected (Table S3). Of these, 464 911, 555 884, 1 146 191, 363 883 and 318 546 SNPs were located in introns, exons, intergenic regions, upstream regions and downstream regions, respectively (Figure S1 and Table S3). The SNP annotation revealed 2,850 splicing, 312 857 nonsynonymous, 1085 stop‐loss and 16 416 stop gain SNPs in the coding sequences (CDS) (Table S3). The chromosomal distribution of SNPs among the 12 rice chromosomes ranged from 334,353 SNPs on Chr. 1 to 192 058 SNPs on Chr. 9 (Table S4). Additionally, SNP frequency was highest on Chr. 8 (8.50 SNPs/kb) (Figure S2 and Table S4).
Figure 1a shows the phylogenetic tree of 259 rice core varieties. A population–structure analysis showed that these 259 rice lines comprised three genetic groups (K = 3): japonica (19), landrace (94) and an improved indica cultivar (146) (Figure 1b and Table S1). A principal component analysis (PCA) showed that the first two PCs explained 29.54% of the genetic variation within these 259 rice lines (Figure 1c). These results indicate that the rice accessions selected in this study represent abundant genetic variation in the core rice germplasm despite some variation due to sequence errors. Based on the r 2 value, which declined to half of the maximum value, the linkage disequilibrium (LD) decay for 259 rice cultivars was estimated at 194 kb (Figure 1d), indicating that the rice lines exhibited a moderate LD (Huang et al., 2010).
Figure 1.
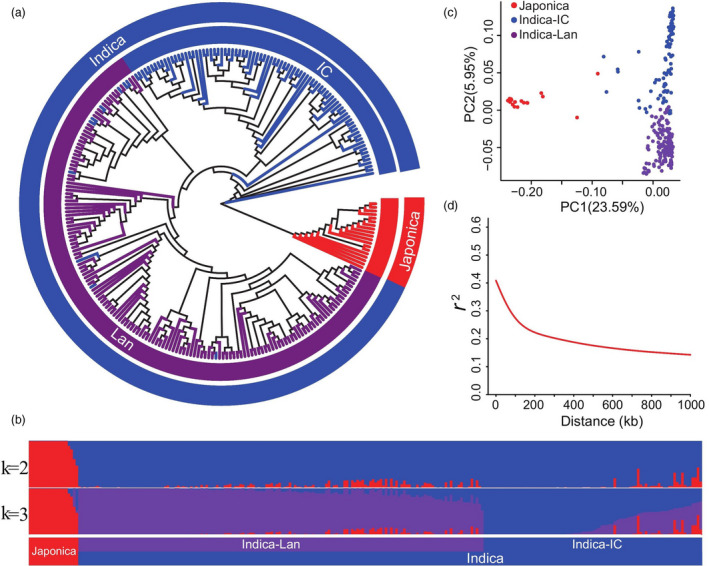
Population structure of 259 rice accessions. (a) Population structure based on different numbers of ancestry kinships (K) set to 2 or 3. The x‐axis indicates the japonica (red), improved indica cultivar (IC, blue), and landrace (Lan, purple) subgroups. The left y‐axis quantifies genetic diversity in each accession. (b) Neighbour‐joining phylogenetic tree of 259 rice accessions based on 2 888 332 high‐quality SNPs; branch colours indicate different subgroups of rice, using the same colours as in (a). (c) Principal component analysis (PCA) plots of the first two components for all 259 rice accessions, using the same colours as in (a). (d) Genome‐wide average linkage disequilibrium (LD) decay rate of 259 rice accessions.
Phenotypic variation among rice cultivars
Rice plants are more susceptible to R. solani at the seedling, tillering and booting stages under high humidity and temperature conditions (Molla et al. 2020; Oreiro et al. 2019). These stages are considered key for R. solani infection because they directly affect rice yield. Therefore, we investigated the resistant phenotypes in different ecological niches in China (Wenjiang and Lingshui) and in different years. At the seedling stage, the RSB incidence rate in all lines ranged from 11.71 to 100% in Wenjiang (2016; average = 34.97%) and 6.18‐100% in Lingshui (2017; average = 35.85%) (Table S1 and Figure S3). Moreover, in Wenjiang and Lingshui, the RSB incidence rate at the tillering (4.04–87.71% and 5.79–76.87%, respectively) and booting (4.91–82.22% and 4.47–84.97%, respectively) stages displayed more than a 16‐ and 12‐fold difference, respectively, between the resistant and susceptible lines. However, the RSB incidence rate in the three stages was normal (Figure S3). These results indicate a broad range of RSB resistance levels among the rice varieties used in the GWAS analysis. An analysis of variance (ANOVA) of the RSB incidence rate at the three developmental stages revealed significant differences among genotypes (Figure S4), implying the presence of dominant RSB resistance loci.
Identification of significant RSB resistance loci via whole‐genome screening
Based on the 2 888 332 high‐confidence SNPs, a GWAS of the three traits (each growth stages was considered to be a different trait) was performed using a mixed linear model (MLM). The phenotypic data of the three traits at two locations were used in the GWAS analysis, as well as the best linear unbiased prediction (BLUP) values for two locations of each trait. A total of 1, 396 SNP loci had a significant association with the three traits (−log10P ≥ 6) (Figure 2a, Figure S5 and Table S5), of which 968 SNP loci were common to at least two traits. Among the three growth stages, the number of associated SNPs was highest at the tillering stage. We further detected candidate genes within 200 kb upstream and downstream of the associated SNP loci, based on the LD decay in the whole genome (Figure 1d). A total of 8, 371 expressed genes were detected among the three traits. The number of these genes was higher on Chr. 3, Chr. 8, Chr. 7 and Chr. 11 than on the other chromosomes (Table S6). Some of the previously reported RSB resistance genes were also included in the GWAS results, including phenylalanine ammonia‐lyase 4 (OsPAL4), loose plant architecture 1 1 (LPA1), oxalate oxidase 4 (OsOXO4), auxin efflux carrier component (OsPIN1a) and chitinase 11 (OsCHI11) (Figure 2c and Figure S6) (Karmakar et al., 2016; Miyashita et al., 2010; Sun et al., 2019; Tonnessen et al., 2015).
Figure 2.
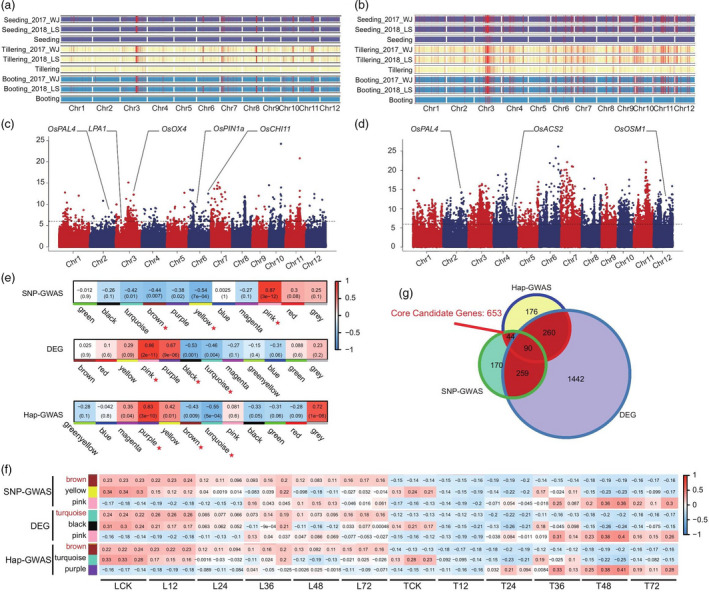
Genome‐wide association study (GWAS) and weighted gene co‐expression network analysis (WGCNA) of rice accessions for identification of rice sheath blight (RSB) resistance candidate genes. (a, b) Distribution of the loci associated with RSB resistance in rice based on single nucleotide polymorphism (SNP)‐GWAS (a) and haplotype (Hap)‐GWAS (b). Incidence of RSB was classified into three stages: seeding (purple); tillering (milky); booting (blue). The associated loci for each trait are indicated by red vertical lines in the chromosome map. Seeding_2017_WJ indicates the GWAS result for RSB resistance at seeding stage using 2017 phenotypic data; Seeding_2018_LS indicates the GWAS result for RSB resistance at seeding stage using 2018 phenotypic data; Seeding indicates the GWAS result for RSB resistance at seeding stage using the best linear unbiased prediction (BLUP) values for two years. The same markers were used in tillering and booting stages. (c, d) Genes with known function for rice disease resistance under candidate genes are shown in significant GWAS association positions, SNP‐GWAS (c), Hap‐GWAS (d). The x‐axis indicates the genomic coordinates, and the y‐axis indicates the association score of each SNP; the score represents a transformed P value, ‐log10P. (e) Module detection for candidates in the GWAS and DEGs data sets. The correlation of different modules is indicated below. We selected one module showing positive correlation with the trait and two modules showing negative correlation with the trait for further study (high expression of resistance genes leads to low RSB incidence thus the negative correlation modules were selected). Each pane corresponds to a module. The module–trait relationships (MTRs) are coloured based on their correlation: red indicates a strong positive correlation and blue indicates a strong negative correlation. (f) Three modules showing the highest correlation with each trait. We selected the module showing clear correlation with the trait as the final candidate module (gene expression was negatively correlated with the resistant phenotype and positively correlated with the susceptible phenotype). Each row corresponds to a module. Every three columns corresponds to a time result. The MTRs are coloured based on their correlation: red indicates a strong positive correlation and blue indicates a strong negative correlation. (g) Venn diagrams showing the number of core candidate genes detected by GWAS and transcriptome analysis.
The traditional SNP‐GWAS method results in a high rate of false‐negative results, and the identification of all genes related to complex traits is difficult because of the presence of a large number of loci with small effects (Ma et al., 2018). To overcome this limitation of SNP‐GWAS, we performed a Hap‐GWAS using 67, 081 haplotype blocks identified from the whole genome in our data. The results showed that 5, 549 haplotype blocks were significantly associated with RSB resistance in the three growth stages (Figure 2b, Figure S7 and Table S7). These haplotype blocks contained 7, 691 potential candidate genes (Table S8). A comparison of the SNP‐GWAS and Hap‐GWAS results revealed 2, 947 candidate genes common to both data sets. However, some previously reported RSB resistance genes were unique to Hap‐GWAS data; for example, haplotype block 66, 367 (Hap‐block ID: 66, 367) on Chr12, was located in the CDS of osmotin 1 (OsOSM1) (Figure 2d and Figure S6), which positively regulates RSB resistance and is mainly expressed in the leaf sheath at the booting stage (Xue et al., 2016).
To further confirm that these candidate genes were associated with RSB resistance, we compared our results with those of previously reported quantitative trait loci (QTLs). Of the candidate genes that were detected in the seedling stage, the number of located on QTL previously identified from biparental mapping studies was 3,175 (SNP‐GWAS) and 3543 (Hap‐GWAS) (Figure S8a). Of the loci that were detected in the tillering and booting stage, the number of located on QTL previous reported ranged from 4566 (Hap‐GWAS) −4846 (SNP‐GWAS) and from 4557 (Hap‐GWAS) −4602 (SNP‐GWAS), respectively (Figure S8b, c). This confirmed that our data were accurate and reliable, indicating a strong potential for the mining novel resistance genes.
Identification of new RSB resistance genes by GWAS and WGCNA
To further identify the core candidate genes associated with RSB resistance, we performed a WGCNA of 8, 371 genes (SNP‐GWAS data; Table S6), 7, 691 genes (Hap‐GWAS data; Table S8) and 13, 524 differentially expressed genes (DEGs) (transcriptome data of the resistant variety Teqing and susceptible variety Lemont, which were inoculated at different time points; Table S9). In the SNP‐GWAS data set, a total of 11 co‐expression modules were detected (Figure S9 and Table S10), with the number of genes in each module ranging from 45 (grey module) to 911 (turquoise module) (Table S10). In the Hap‐GWAS data set, 12 co‐expression modules were detected (Figure S9 and Table S11), with each module comprising 74 (greenyellow module) to 726 (turquoise module) genes (Table S11). Additionally, 13, 524 DEGs were assigned into 12 co‐expression modules (Figure S9), each comprising 22 (grey module) to 2051 (turquoise module) genes (Table S12). To identify the modules associated with RSB resistance, we correlated the selected modules with RSB resistance and searched for significant associations. The results showed that three modules each in the SNP‐GWAS data (pink, r = 0.87, P = 3e−12; yellow, r = −0.54, P = 7e−04; and brown, r = −0.44, P = 0.007), Hap‐GWAS data (purple, r = 0.83, P = 3e−10; turquoise, r = −0.55, P = 5e−04; and brown, r = −0.43, P = 0.009) and DEGs data (pink, r = 0.86, P = 2e−11; black, r = −0.53, P = 0.001; turquoise, r = −0.46, P = 0.004) had a significant correlation with RSB resistance (Figure 2e). Among these, SNP‐GWAS candidate genes in the brown module (n = 563), Hap‐GWAS candidate genes in the brown module (n = 570) and DEGs in the turquoise module (n = 2, 051) had the strongest positive correlation with RSB incidence in the susceptible genotype Lemont, and the highest negative correlation in the resistant genotype Teqing (Figure 2f and Table S13). Interestingly, in these three modules, 653 genes were found to be shared by at least two modules and were named ‘core candidate genes’ (Figure 2g and Table S14).
Moreover, these 653 genes were categorized into two different expression profiles: (Ⅰ) a total of 537 genes (named type 1), which had a high expression level in the susceptible variety Lemont after inoculation with R. solani AG1‐IA, and a low expression level in the resistant variety Teqing; and (Ⅱ) a total of 116 genes (named type 2), which had a high expression level in the resistant variety Teqing after inoculation with R. solani AG1‐IA, and a low expression level in the susceptible variety Lemont (Figure S10). Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses showed that type 2 genes were associated with kinase activity; positive regulation of developmental process; cutin, suberin and wax biosynthesis; glutathione metabolism; and fatty acid degradation (Figure S10), and these terms or pathways were involved in plant resistance. A total of 71 genes were detected among the type 2 gene set after the removal of genes encoding unknown/transposon/retrotransposon proteins, and another eight genes had been previously published (Table S14). These 71 genes were therefore selected for further analysis.
OsRSR1 positively regulates resistance to R. solani AG1‐IA
NLR genes play a vital role in the plant response to pathogenic bacteria (Marone et al., 2013). Among the 71 type 2 genes, there were three NLR genes: LOC_Os11g12320, LOC_Os11g12330 and LOC_Os11g12340 (Table S14). We further found that these three genes and LOC_Os11g12350 formed an NLR gene cluster (R‐cluster), which showed a significant association with RSB resistance (−log10P = 6.269), based on SNP‐GWAS results (Figure S11). This R‐cluster mapped to a 36.83 kb size (6 876 516–691 3345 bp) on Chr. 11 (Figure 3a) and formed a co‐localized LD block (block 56755) with a significant association with SNP 6898599 (A/G) (Figure 3b). We further analysed the R‐cluster sequences to identify structural variations and discovered a 21.8 kb deletion in the sequences. These structural variations formed three haplotype groups (Figure 3c), and rice accessions carrying the deletion haplotype (hap1) were more susceptible than those of the hap2 and hap3 (Figure 3d). Transcriptome data showed that the expression of LOC_Os11g12320, LOC_Os11g12330 and LOC_Os11g12340 in the resistant variety Teqing was much higher than that in the susceptible variety Lemont after inoculation with R. solani AG1‐IA (Figure 3e); however, the most significant difference was in the expression of LOC_Os11g12340 (named OsRSR1) in the resistant and susceptible variety. These results were verified by a real‐time quantitative polymerase chain reaction (qRT‐PCR) (Figure 3f). Furthermore, the PCR amplification results showed that OsRSR1 was absent in seven randomly selected rice lines (with the deletion haplotype), which verified that the deletion haplotype was accurate (Figure S12). A homogeneous analysis demonstrated that OsRSR1 was found to share a close genetic relationship with AT3G07040.1 (Figure S13), which encodes the NB‐ARC domain‐containing disease resistance protein and confers resistance to P. syringae, in Arabidopsis (Jung et al., 2020). These results indicate that this R‐cluster was significantly associated with RSB resistance and the OsRSR1 could be a candidate sheath blight resistance gene.
Figure 3.
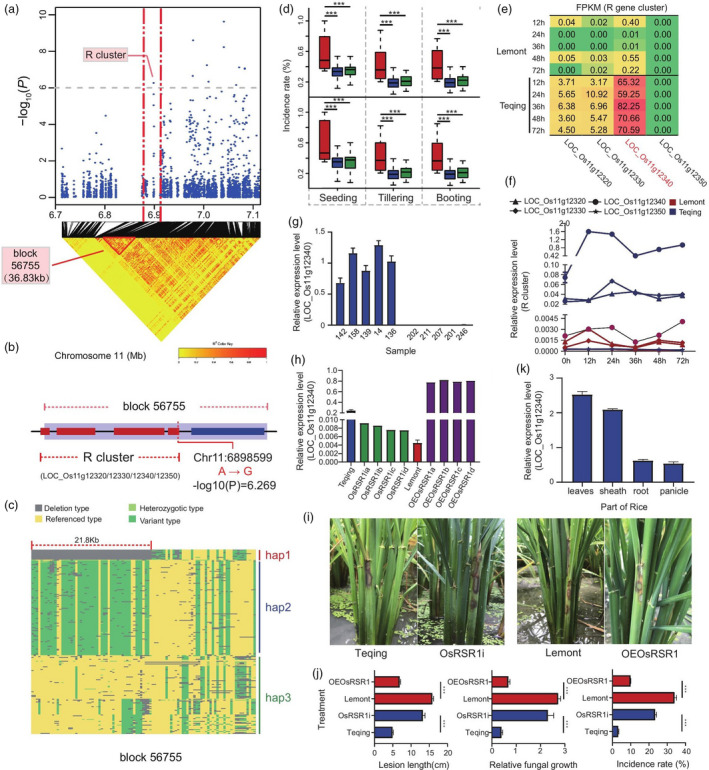
OsRSR1 regulates RSB resistance. (a) Manhattan plots of loci on chromosome 11 associated with RSB incidence at the tillering stage at Wenjiang in 2017 (Tillering_2017_WJ). Arrowheads indicate significantly associated SNPs located in a nucleotide‐binding site leucine‐rich repeat proteins encoded (R) gene cluster. Horizontal dashed lines indicate the significance threshold (P < 10−6). LD heat map (bottom) reflected that associated SNP localized in a haploid between the red dashed lines. (b) This haploid (named block 56 755) contains the R gene cluster mentioned above, which included four R genes LOC_Os11g12320, LOC_Os11g12330, LOC_Os11g12340 and LOC_Os11g12350. Red rectangles indicate four R genes, respectively. (c) Three haplotypes were identified through sequence analysis of R gene cluster. Blue areas indicated deletion type mapping to reference genomes (hap1); green areas indicated variant type mapping to reference genomes (hap2); yellow areas indicated consistent with reference genomes (hap3). (d) Box plots for RSB resistance, based on the genotypes of sequence analysis of R cluster. The horizontal line in the centre of each box denotes the median. The upper and lower limits of each box represent quartiles; whiskers indicate the range of the data; statistical significance of differences was analysed by the two‐tailed t‐test. Hap1, hap2 and hap3 are shown in red, blue and green, respectively. (e) Expression of four genes in the R gene cluster in resistant and susceptible rice lines (Transcriptome data). Each row corresponds to a time result. Each column corresponds to a gene. The expression level is coloured based on their FPKM value: red indicates a high expression and green indicates a low expression. (f) Verification of four R genes expression in resistant and susceptible rice lines at different infection time points by qRT‐PCR. (g) Expression analysis of OsRSR1 in five resistant and five susceptible varieties at 24 h postinoculation (hpi) of Rhizoctonia solani AG1‐IA by qRT–PCR. (h) Identification of overexpression (OE) and RNA interference (RNAi) lines of OsRSR1 by qRT–PCR. (i) Incidence of OsRSR1‐OE plants was decreased compared with WT, and the incidence of OsRSR1‐RNAi plants was increased compared with WT. (j) Average incidence rate (n = 10 sheath) of Teqing, OsRSR1‐RNAi lines, Lemont, and OsRSR1‐OE lines at 4 day post inoculation (dpi) with R. solani AG1‐IA. (k) Expression analysis of OsRSR1 in different tissues of rice by qRT–PCR (leaves, sheath, root and panicle). The rice UBQ gene was used as an internal control. Data are represented as average values with four biological replicates (e–h, and k).
To confirm this result, we randomly selected five resistant and five susceptible varieties to examine the expression level of OsRSR1 after inoculation with R. solani AG1‐IA. Expression of the OsRSR1 gene was higher in resistant varieties than that in susceptible varieties at 24‐h postinoculation (hpi) (Figure 3g). We then investigated the function of OsRSR1 in rice using the transgenic approach. The expression of OsRSR1 was either down‐regulated by RNA interference (RNAi) or enhanced by expressing OsRSR1 under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The expression of OsRSR1 in OsRSR1‐RNAi and overexpression (OE) lines was verified by qRT‐PCR (Figure 3h). The expression of OsRSR1 in OsRSR1‐OE lines was 20‐fold greater than in the wild‐type (WT) (Figure 3h). The T1 progenies of the OsRSR1 RNAi and OE lines were evaluated for disease resistance and lesion length and mycelia biomass was measured at 4 days postinoculation (dpi). The OsRSR1‐RNAi plants of the resistant genotype Teqing were more susceptible than the WT (Figure 3i, j and Figure S14). We then transformed the susceptible rice variety Lemont with the dominant OsRSR1 allele of Teqing, and the resulting transgenic lines exhibited a significantly enhanced resistance to R. solani AG1‐IA compared to the WT (Figure 3i, j and Figure S14). Additionally, the expression of OsRSR1 was higher in leaves and leaf sheaths, which characteristically show symptoms of sheath blight, than in roots and panicles (Figure 3k). Moreover, there were no significant differences in yield and agronomic traits between OsRSR1‐OE and WT plants (Figure S15). In a further examination, the T2 progenies of OsRSR1 RNAi and OE lines were evaluated for disease resistance in the field and were found to exhibit similar results to those obtained for T1 (Figure S16a, b). These results suggested that OsRSR1 positively regulates resistance to R. solani AG1‐IA without compromising fitness.
OsRLCK5 positively regulates resistance to R. solani AG1‐IA
The enrichment of kinase activity in the type 2 gene set indicated that protein kinase plays a key role in the regulation of RSB resistance. Therefore, we also analysed LOC_Os01g02390 (named OsRLCK5), which encodes the protein kinase domain‐containing protein, and was associated with RSB resistance, based on the Hap‐GWAS results (Figure 4a and Figure S17). Haplotype block 70 on Chr. 1 was located in the CDS of OsRLCK5 (Figure 4a, b). There were five subtypes in this haplotype: GTAG, ATAG, AGAG, AGGG and AGGA, and AGGA showed as significant association with RSB resistance (−log10P > 6 in the four traits association results) (Figure 4b). One synonymous SNP, Chr1_769280 (A/G), and three nonsynonymous SNPs, Chr1_769292 (G/T), Chr1_769480 (G/A) and Chr1_769492 (A/G) in OsRLCK5, were formed in this haplotype (Figure 4b, c). We analysed two homozygous haplotypes (AGGG and AGGA) and found that accessions carrying the haplotype AGGG were resistant, whereas those with the haplotype AGGA were susceptible (Figure 4d). Furthermore, these findings were confirmed by the genotyping results of the nonsynonymous SNP Chr1_769492 (A/G), which indicated that accessions carrying the homozygous GG allele were more resistant than those with the heterozygous AG and homozygous AA alleles (Figure 4d). In addition, transcriptome data showed that the expression of OsRLCK5 dramatically increased in Teqing at 12 h and remained high throughout the experiment, but there was almost no expression in Lemont (Figure 4e). The qRT‐PCR results were consistent with the transcriptome data (Figure 4f). Furthermore, in Teqing, the expression of OsRLCK5 in the leaf and leaf sheath was higher than in the panicle and root at 24 hpi (Figure 4g). Additionally, there was a higher expression in OsRLCK5 in five randomly selected resistant lines than in susceptible lines at 24 hpi (Figure 4h). A comparison of its kinase domain with others from the blast results revealed that several typical conserved serine‐threonine protein kinase subdomains were found in OsRLCK5 (Figure S18), and some homogeneous genes of OsRLCK5 were found in Arabidopsis (Figure S13). These results suggest that the OsRLCK5 protein is a typical serine‐threonine kinase, and the expression of OsRLCK5 may be induced following R. solani AG1‐IA infection, enabling it to participate in RSB resistance.
Figure 4.
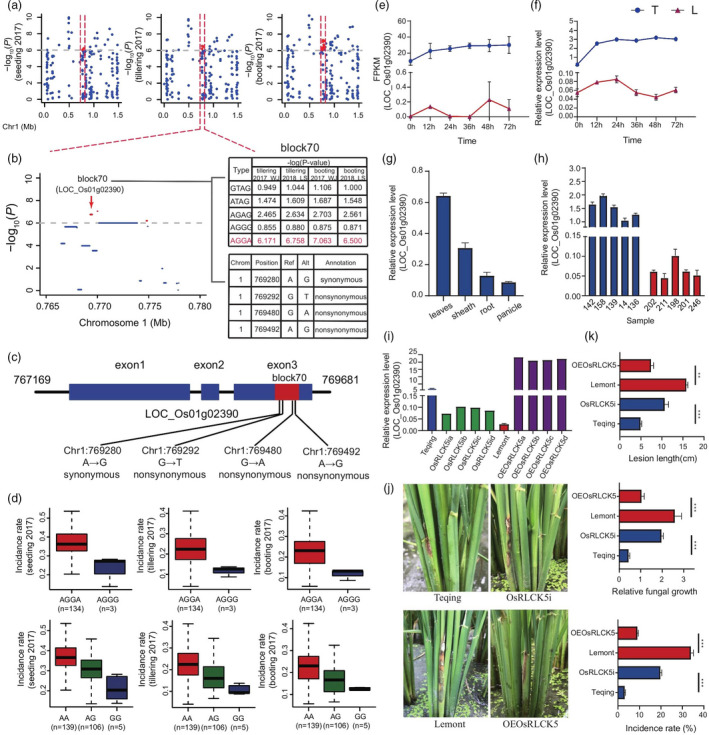
OsRLCK5 regulates RSB resistance. (a) Manhattan plots of loci on chromosome 1 associated with RSB incidence at the seedling, tillering and booting stages at Wenjiang in 2017 (seedling_2017_WJ, tillering_2017_WJ and booting_2017_WJ, respectively). Red markings indicate the strongly associated loci containing the candidate gene OsRLCK5. Horizontal dashed lines indicate the significance threshold (P < 10−6). (b) Local Manhattan plot. The candidate region lies between the red dashed lines. Significant haplotype structures (block 70), P‐values and variant types are indicated on the right side. Dashed line represents the significance threshold (P < 10−6). Red indicated the strongly associated haplotype, which are located within OsRLCK5. (c) Haplotype with three nonsynonymous variations in OsRLCK5 exon. Blue rectangles and black lines indicate exons and introns, respectively. (d) Box plots for RSB resistance, based on the genotypes of two homozygous haplotypes. The horizontal line in the centre of each box denotes the median. The upper and lower limits of each box represent quartiles; whiskers indicate the range of the data. n indicates the number of accessions with the same genotype. (e) Expression analysis of OsRLCK5 in five resistant and five susceptible varieties at 24 hpi by qRT–PCR. (f) Verification of OsRLCK5 expression in resistant and susceptible rice lines at different infection time points by qRT‐PCR. (g) Spatial expression analysis of OsRLCK5 in rice plants. (h) Expression analysis of OsRLCK5 in five resistant and five susceptible varieties at 24h postinoculation (hpi) of R. solani by qRT–PCR. (i) Identification of OsRLCK5‐OE and ‐RNAi lines by qRT–PCR. The rice UBQ gene was used as an internal control. Data are represented as average values with four biological replicates (e–i). (j) Incidence of OsRLCK5‐OE plants was decreased compared with WT, and the incidence of OsRLCK5‐RNAi plants was increased compared with WT. (k) Average incidence rate (n = 10 sheath) of Teqing, OsRLCK5‐RNAi lines, Lemont and OsRLCK5‐OE lines at 4 dpi with R. solani AG1‐IA.
To further verify our results, we generated OsRLCK5‐RNAi lines in a Teqing background and OsRLCK5‐OE lines in a Lemont background. Transcript levels of OsRLCK5 were substantially reduced in OsRLCK5‐RNAi lines and markedly elevated in OsRLCK5‐OE lines compared with Lemont (Figure 4i). Inoculation of transgenic T1 lines with R. solani AG1‐IA revealed enhanced symptoms (lesion length and mycelial biomass) in OsRLCK5‐RNAi plants compared with the WT (Figure 4j, k and Figure S19) and significantly enhanced resistance in OsRLCK5‐OE lines compared with Lemont at 4 dpi (Figure 4i, k and Figure S19). Moreover, there were no significant differences in the yield and agronomic traits of OsRLCK5‐OE lines compared with the WT (Figure S15). The transgenic T2 lines were evaluated for disease resistance in the field and the results were similar to those obtained for T1 (Figure S16a, b). These results suggest that OsRLCK5 positively regulates resistance to R. solani AG1‐IA without a fitness cost.
OsRSR1 and OsRLCK5 enhance defence activation
We compared the expression of JA (OsAOC, OsAOS2) (Mei et al., 2006; Michael et al., 2013), ET (OsACS2) (Iwai et al., 2006) and SA (OsNPR1) (Li et al., 2013) signalling genes in WT, RNAi and OE plants at 72 hpi. The expression levels of OsAOC, OsAOS2 and OsACS2 were significantly higher in OsRSR1‐OE lines than in Lemont and significantly lower in OsRSR1‐RNAi plants than in Teqing (Figure S20). However, there was no significant difference in the expression of OsNPR1, a marker gene of the SA‐induced resistance response, between OsRSR1 transgenic plants and the WT (Figure S20). The expression levels of OsAOC, OsAOS2 and OsACS2 were significantly enhanced in OsRLCK5‐OE lines compared with Lemont and down‐regulated in OsRLCK5‐RNAi lines compared with Teqing after AG1‐IA infection (Figure S20). Furthermore, the qRT‐PCR analysis showed that at 72 h, the transcript levels of PR1b and PAL were significantly higher in OsRSR1‐OE lines than in Lemont; however, the transcript level of PR1b, PR10a, and PAL was down‐regulated in OsRSR1‐RNAi lines compared with Teqing plants. Similarly, the expression levels of PR1b, PR10a and PAL in OsRLCK5‐OE plants were significantly higher than in Lemont (Figure S20), while only PAL had a significantly lower expression in OsRLCK5‐RNAi lines compared with Teqing (Figure S20). Interestingly, OsACO2 and OsACO3 are key genes in ET‐induced plant defence, and OsLOX4 and OsLOX1, which are important JA biosynthesis genes, were identified in the core candidate gene set (Table S6).
OsRSR1 and OsRLCK5 are involved in the ROS pathway to resistance
The N‐terminal coiled‐coil (CC) domain of some NLR proteins is required for interaction with downstream signalling components or for the induction of immune responses (Shen et al., 2007). We therefore performed yeast two‐hybrid (Y2H) assays using the CC domain of OsRSR1 (OsRSR1‐CC; 3–133 aa) as bait. From a complete rice cDNA library, we identified several OsRSR1‐CC interactors, including an OsSHM1 (Figure 5a) and a serine hydroxyl methyltransferase that was involved in photorespiration and ROS generation (Wang et al., 2015), but had not previously been reported to have a role in plant disease resistance. We further confirmed the interaction between OsRSR1‐CC and full‐length OsSHM1 using bimolecular fluorescence complementation (BiFC) assays. Fluorescence signals were mainly distributed in the cytoplasm and nucleus (Figure 5b); thus, we speculated that OsSHM1 is involved in OsRSR1‐mediated RSB resistance. We also identified the OsRLCK5 interaction partner using the Y2H assay, a glutaredoxin (OsGRX20), which is involved in ROS regulation (Rohini et al., 2010; Ströher and Millar, 2012) and was identified among 69 potential interactors. The direct interaction between OsRLCK5 and OsGRX20 was confirmed via a BiFC analysis by their co‐expression in tobacco leaf epidermal cells (Figure 5a, b). Based on these results, we speculated that OsRSR1 and OsRLCK5 confer RSB resistance by regulating ROS.
Figure 5.
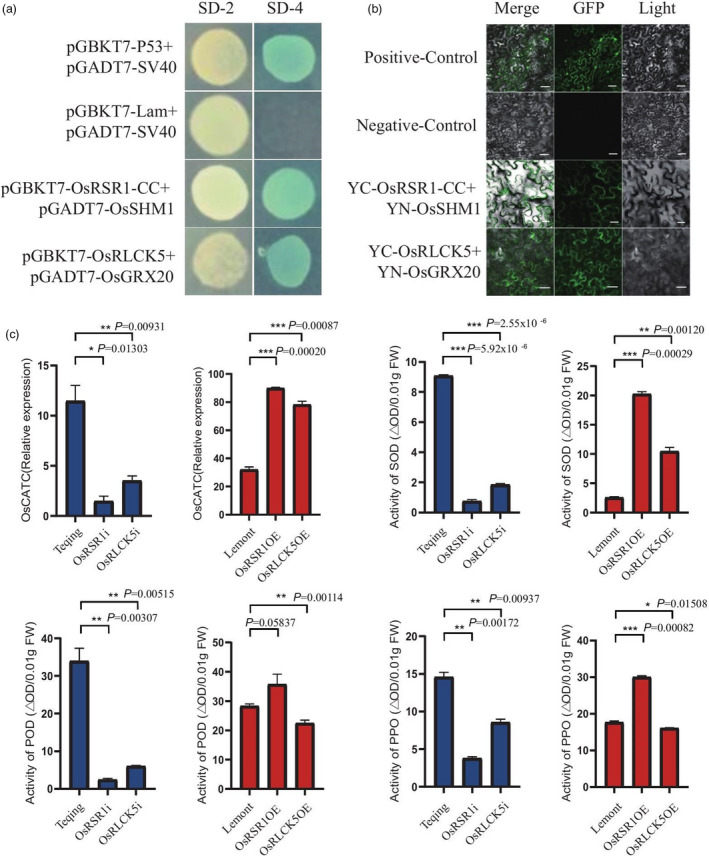
OsRSR1 and OsRLCK5 regulate reactive oxygen species (ROS) burst. (a) Detection of interaction between OsRLCK5 and OsGRX20, and OsRSR1‐CC and OsSHM1 in a yeast two‐hybrid (Y2H) assay, respectively. Photograph shows the growth behaviour of transformants on SD/Leu‐Trp media (SD‐2) and SD/Ade‐Leu‐Trp‐His (SD‐4) plus AbA and X‐α‐gal media. pGBKT7‐p53 and pGADT7‐SV40 large T‐antigen were set as the positive control pair. pGBKT7‐Lam and pGADT7‐SV40 large T‐antigen were set as the negative control pair. (b) Bimolecular fluorescence complementation (BiFC) assay verified the interaction between OsRLCK5 and OsGRX20, and OsRSR1‐CC and OsSHM1 in tobacco leaf epidermis cells, respectively. Scale bars: 50 mm. 35S‐YFP was employed as positive control, and negative control means the pXY104 (nYFP; contains OsRLCK5 or OsRSR1‐CC) were co‐expressed in tobacco leaf epidermis cells with empty pXY106 (nYFP) vector. (c) Changes in ROS levels, activities of POD, SOD and PPO, and OsCATC expression in transgenic and wild type (WT; Teqing and Lemont) leaves at 72 hpi. Values of POD, SOD and PPO activities represent the average of four lines. Data indicate mean ± standard error of mean (SEM) of four technical replicates. Statistically significant differences were analysed by one‐way ANOVA (*P < 0.05; **P < 0.01; ***P < 0.001).
To further confirm our speculation, we examined the activity of ROS‐related antioxidant enzymes and the relative expression of OsCATC in transgenic and WT rice plants. The relative expression of OsCATC in OsRSR1‐RNAi was significantly reduced compared with Teqing, and significantly increased in OsRSR1‐OE plants compared with Lemont at 72 hpi (Figure 5c). Moreover, the peroxidase (POD), superoxide dismutase (SOD) and polyphenol oxidase (PPO) activities were significantly reduced in OsRSR1‐RNAi plants compared with Teqing (Figure 5c), while the SOD and PPO activities were significantly increased in OsRSR1‐OE plants compared with the WT; however, there were no significant differences in POD activity between OsRSR1‐OE plants and Lemont (Figure 5b). Similar results were obtained in OsRLCK5 transgenic lines (Figure 5c). Additionally, following R. solani AG1‐IA infection, more DAB‐ and NBT‐stained spots appeared on the leaves surrounding the lesions on OsRSR1‐OE plants than on the WT (Figure S21), suggesting that OsRSR1‐OE plants increased the accumulation of H2O2 and O2− in the transgenic rice. These data suggested that OsRSR1 and OsRLCK5 positively regulate the transcription of OsCATC and influence the SOD and PPO activities, thus affecting the ROS content of rice plants infected by R. solani AG1‐IA.
The GSH‐AsA antioxidant system was activated in the overexpression of OsRSR1 and OsRLCK5
The interaction between 653 core candidate genes was performed through a protein–protein interaction network analysis (Figure S22). We found an interaction between some proteins that are involved in the GSH‐AsA antioxidant system (a nonenzyme system involved in scavenging ROS, Conklin, 2001; Sun et al., 2018). For example, there were interactions between cytosolic dehydroascorbate reductase (OsDHAR1, Kim et al., 2013) and lipoxygenase 1 (OsLOX1), the glutathione synthetase gene LOC_Os11g42350, the SOD gene LOC_Os06g02500, the glutathione peroxidase gene LOC_Os11g18170, and the glutathione S‐transferase gene LOC_Os01g72170. Thus, we speculated that the GSH‐AsA antioxidant system may be involved in RSB resistance and was mediated by OsRSR1 and OsRLCK5.
We further examined the AsA, dehydroascorbate (DHA), GSH, and oxidized glutathione (GSSG) content in transgenic and WT rice plants at 72 hpi. The AsA and DHA levels in OsRSR1‐OE plants were significantly enhanced compared with Lemont, while no significant difference was found between OsRSR1‐RNAi and Teqing plants at 72 hpi. Consistent levels of GSH and GSSG were found, with the contents of these two enzymes being clearly higher in OsRSR1‐OE than in Lemont plants at 72 hpi (Figure S23). For OsRLCK5 transgenic lines, only the levels of GSH and GSSG were significantly different between transgenic and WT plants at 72 hpi (Figure S23). The GSH and GSSG contents in OsRLCK5‐RNAi were significantly reduced compared with Teqing and significantly increased in OsRLCK5‐OE plants compared with Lemont at 72 hpi (Figure S23). We also analysed the expression levels of OsAPX1, DHAR1 and GRX20 in transgenic and WT rice plants at 72 hpi. These genes play key roles in the AsA‐GSH antioxidant system. The relative expression of OsAPX1, DHAR1 and GRX20 in OsRSR1‐OE was significantly increased compared with Lemont and significantly reduced in OsRSR1‐RNAi plants compared with Teqing at 72 hpi (Figure S24). A similar expression pattern was found in OsRLCK5 transgenic lines (Figure S24). These results indicated that the AsA‐GSH antioxidant system was involved in the process of resistance to R. solani AG1‐IA regulated by OsRSR1 and OsRLCK5.
Discussion
RSB caused by R. solani AG1‐IA is one of the most destructive diseases in rice (Srinivasachary and Savary, 2011). Mining gene resource that is resistant to RSB, and applying elite resistance genes to improve the resistance of rice is the most practical and effective way to manage the disease. The analysis and mapping of RSB resistance genes using traditional genetic methods are difficult because inoculation methods and environmental factors affect the accuracy of RSB resistance phenotyping. For these reasons, most previous studies of QTL mapping for RSB resistance in rice have been based on early‐segregating mapping populations that could not be evaluated repeatedly (Zuo et al., 2013; Zuo et al., 2007; Zou et al., 2000; Wang et al., 2002). Recently, RSB studies that have utilized GWAS approaches have reported a number of QTLs contributing to resistance (Chen et al., 2019; Oreiro et al., 2019). However, these reports have had limited success in the identification of functional genes because a large number of loci are associated with traits. In this study, we obtained 653 core candidate genes associated with RSB resistance using a combination of SNP‐GWAS, Hap‐GWAS and WGCNA. A WGCNA can overcome the limitations of a GWAS by identifying the functional connections among genes in an unbiased manner using trait‐relevant expression data (Liu et al., 2017; Van‐Nas et al., 2009).
Although substantial progress has been made in the mapping and cloning of resistance genes or QTLs, only a few resistance genes against R. solani AG1‐IA have been cloned in rice. For example, WRKY30 positively regulates RSB resistance by activating JA biosynthesis‐related genes and the subsequent increase of endogenous JA accumulation (Peng et al., 2012). Both WRKY4 and WRKY80 have been found to enhance RSB tolerance by inducing the up‐regulation expression of JA‐ and ET‐responsive PR genes (Peng et al., 2016). The study reported that upon RSB infection, the rice polygalacturonase inhibiting protein OsPGIP1 can be induced, and improving the RSB tolerance in transgenic rice plants overexpressing the OsPGIP1 gene (Chen et al., 2016). Although these genes have been detected, little progress has been made in rice breeding for RSB resistance, with none of these genes selected for use in breeding RSB resistance. In this study, the ability of OsRSR1 and OsRLCK5 to confer RSB resistance in rice was identified by adopting a transgenic approach. These two loci were the first cloned RSB‐related QTL and therefore represent a crucial target for developing R. solani AG1‐IA resistance through rice breeding. Furthermore, the haplotype analysis of OsRLCK5 revealed the presence of the resistant allele (GG) in six improved cultivars. This is probably because the OsRLCK5 allele is the result of natural selection and breeding. However, because this allele was identified in only 4.3% of our selected improved rice cultivars, there is a large potential for the introduction of this natural allele into other rice varieties. Additionally, OsRSR1 and OsRLCK5 transgenic plants represent a valuable genetic resource for breeding RSB resistance in rice. More importantly, we obtained transgenic lines showing increased expression of cytochrome P450 LOC_Os07g33440 (CYP716A16) and SAM‐dependent carboxyl methyltransferase LOC_Os06g20960 (OsSam12) in a Nipponbare rice variety background (Figure S25a and Figure S26a). These two genes were selected from the 653 core candidate gene set. Inoculation of transgenic T1 lines with R. solani AG1‐IA resulted in enhanced resistance in OE lines to R. solani AG1‐IA compared with the WT (Figure S25b, c and Figure S26b, c). Thus, our studies provide a valuable genetic resource for the study of RSB resistance; more work is needed to validate additional genes involved in RSB resistance.
Our results demonstrated that OsRSR1‐CC and OsRLCK5 interact with OsSHM1 and OsGRX20, respectively, to regulate the level of ROS, which serve as a secondary messenger in signal transduction, leading to the activation of PR genes and an immune response. Furthermore, the GSG‐AsA antioxidant system played a role in the resistance to R. solani AG1‐IA regulated by OsRSR1 and OsRLCK5. Thus, we developed a model of RSB resistance regulation by OsRSR1 and OsRLCK5 to summarize our results (Figure 6). The overexpression of OsRSR1 results in H2O2 accumulation at the infection site and, more importantly, increased antioxidant activities through activation of the GSG‐AsA antioxidant system. Additionally, OsRLCK5 is involved in the activation of the GSG‐AsA antioxidant system through its interaction with GRX20. We have uncovered a molecular mechanism of RSB resistance mediated by OsRSR1 and OsRLCK5 through their regulation of the level of ROS. Similar results were previously obtained in tobacco, where the suppression of tobacco catalase 1 or catalase 2 resulted in the accumulation of H2O2, elevated levels of PR1 protein and SA, and enhanced resistance to the tobacco mosaic virus (Takahashi et al., 1997).
Figure 6.
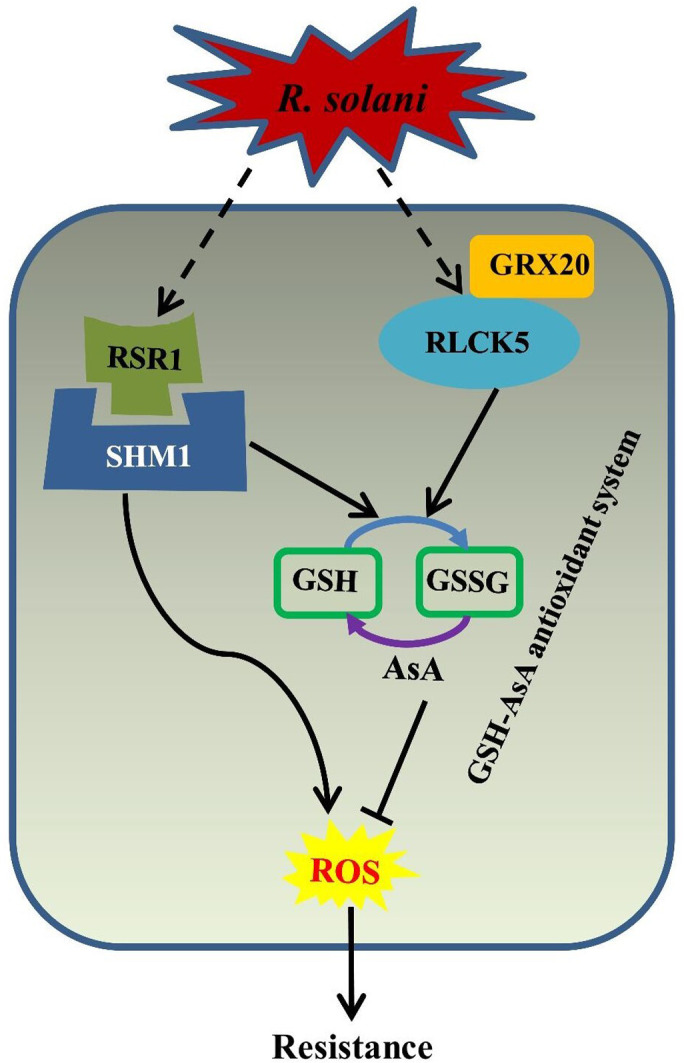
A model for RSR1‐ and RLCK5‐mediated RSB resistance through glutathione (GSH)‐ascorbic acid (AsA) antioxidant system.
We have reported the role of the NLR protein OsRSR1, and RLCK protein OsRLCK5 in RSB resistance for the first time. There were no significant differences in yield and agronomical traits of transgenic OsRSR1 and OsRLCK5 plants compared with WT plants. We first demonstrated that OsRSR1 and OsRLCK5 conferred RSB resistance through the GSG‐AsA antioxidant system. To better understand the molecular mechanisms controlling the OsRSR1 and OsRLCK5 regulated resistance to R. solani AG1‐IA, it is necessary to determine the ability of OsRSR1 and OsRLCK5 to recognize an effector in a pathogen. Further studies are needed to explore the mechanism of disease resistance in rice and to advance our understanding of the resistance pathways involved in the rice–R. solani AG1–IA interaction.
Methods
Pathogen inoculation and disease resistance assay
All 259 rice accessions were evaluated for RSB resistance at the seedling stage using the ‘mist‐chamber’ method, as described previously (Zuo et al., 2007); the experiment was performed under field conditions. In the field, seedling–breeding plates (55 cm long × 29 cm wide × 12 cm deep) with 32 holes were filled with presterilized soil. Six seeds were sown in each hole. Five replicates were prepared for each variety, and plates were placed under natural light conditions. Seedlings were thinned at the third‐leaf stage to retain one uniform seedling per hole. At the fourth leaf stage, the plates were moved into a prebuilt ‘mist‐chamber’ and acclimated for 24 h before pathogen inoculation, which was performed as described previously (Liu et al., 2009), with a slight modification. A standard strain of R. solani AG1‐IA was used for inoculation of rice plants. The pathogen was grown on truncated thin matchsticks (0.8–1.0 cm long × 2–3 mm wide × 1 mm thick) on potato dextrose broth medium at 28 ºC in the dark for 2–3 days. To perform the inoculation, the inoculum was closely affixed to one side of the base of the seedling stem, ensuring that a hypha was directly touching the plant. Five plants per variety were inoculated as replications. When disease symptoms appeared on the whole stems or leaves of the susceptible control Lemont, all 259 rice lines were screened for disease. At the tillering and booting stages, inoculum was placed at the base of the leaf closest to the ground. Fifteen tillers per plant of each variety were used as replications. When the lesion length of the susceptible control Lemont reached approximately 50% of the collar height, the 259 rice lines were screened for morbidity. The RSB disease score was recorded for each seedling as the lesion length divided by the collar height (distance from the ground to the tallest leaf collar on the main stem) (Lavale et al., 2018). Data were processed with Microsoft Excel 2010. A statistical analysis of the RSB scores among different varieties or subpopulations was performed through an analysis of variance (ANOVA) and Dunnett’s multicomparison tests using SPSS version 16.0 (IBM Corp., Armonk, NY).
Other methods
Details of the plant materials and growth conditions, DNA extraction, PCA, population genetics, LD analysis, GWAS and WGCNA analyses, GO and KEGG pathway analyses, plasmid construction, plant transformation, gene expression and antioxidant enzyme activity analysis, Y2H assays, and BiFC assay are provided in the supplementary methods. All primers used for plasmid construction and gene expression analysis are listed in Table S15.
Conflict of interest
The authors declare no competing interests.
Author contributions
AZ and AW designed the project. AW, XS, LC, JZ, LM and YJ performed the experiments. XJ, YN and CJ performed bioinformatics analysis. AW wrote the manuscript. SL, QD, SW, JZ, YL, TZ, HL, LW, YH and PL provided useful advice. All authors read and approved the final manuscript.
Supporting information
Figure S1 The geographic distribution of 259 rice accessions. Each dot of a given colour on the world map represents the geographic distribution of the corresponding rice accession
Figure S2 Distribution of single nucleotide polymorphisms (SNPs) and nucleotide diversity across the rice Nipponbare genome in the rice association panel
Figure S3 The frequency distribution of incidence rate of 259 rice lines after inoculation with Rhizoctonia solani AG1‐IA
Figure S4 Histograms and box plots showing the resistance phenotypic data of 259 rice lines at three grown stages to rice sheath blight (RSB)
Figure S5 Manhattan plots and QQ plots resulting from the SNP‐based GWAS for RSB resistance
Figure S6 Several known rice sheath blast resistant genes were identified by SNP‐based and haplotype‐based GWAS
Figure S7 Manhattan plots resulting from the haplotype‐based GWAS for RSB resistance
Figure S8 Chromosomal positions of loci for RSB resistance identified via SNP‐based and haplotype‐based GWAS. (a) Seedling stage. (b) Tillering stage. (c) Booting stage. The mapped loci in this study are shown on the right, and previously mapped QTLs are shown in pink region
Figure S9 WGCNA of genes in SNP‐GWAS, haplotype‐based GWAS and DEGs three datasets
Figure S10 The expression profiles and pathway analysis of 653 core candidate RSB resistant genes in this study
Figure S11 Manhattan plots for RSB resistant on chromosome 11
Figure S12 PCR amplification results of OsRSR1 in resistant and susceptible rice line. M: DNA marker; 1‐14 represent ID of rice lines 267, 136, 139, 142, 158, 178, 14, 283, 202, 307, 197, 199, 208, and 323, respectively
Figure S13 Phylogenetic and structural analysis of two candidate resistant genes. (a) Phylogenetic analysis of the deduced amino acid sequence of OsRSR1 and OsRLCK5 compared with Arabidopsis thaliana and rice sequences. (b) Schematic diagram of the OsRSR1 and OsRLCK5 proteins
Figure S14 Changes in the disease resistance of OsRSR1‐RNAi and OsRSR1‐overexpressing (OE) transgenic rice plants. Leaf lesion represents the means of four lines. Statistically significant differences were analysed based on four technical replications (one‐way ANOVA; ***P < 0.001). Bars indicate standard errors of the mean
Figure S15 The plant height, number of tillers, setting percentage, and 1,000‐grain weight of OE transgenic plant and wild type (WT) were observed
Figure S16 (a) Expression analysis of OsRLCK5 and OsRSR1 in WT and OE transgenic T2 rice plants by qRT–PCR. (b) Changes in the disease resistance of OsRSR1 and OsRLCK5 T2 transgenic lines. Leaf lesion represents the means of five lines. Statistically significant differences were analysed based on four technical replications (one‐way ANOVA; **P < 0.01, ***P < 0.001). Bars indicate standard errors of the mean
Figure S17 Manhattan plots for RSB resistant on chromosome 01
Figure S18 Alignment of the conserved serine‐threonine domain of OsRLCK5, its homologs in Arabidopsis thaliana, and other rice serine‐threonine kinases. Underlined regions represent the conserved subdomains of serine/threonine protein kinases; the residues marked with asterisks are the three phosphorylated catalytic sites
Figure S19 Changes in the disease resistance of OsRLCK5‐RNAi and OsRLCK5‐OE transgenic rice plants. (a) Disease symptoms in TeQing, Lemont, OsRLCK5‐RNAi and OsRLCK5‐OE plants at 72 h after inoculation with AG1 IA. Leaf lesion represents the means of four lines. (b) Disease severity was evaluated as relative lesion area at 72 hour postinoculation (hpi). Statistically significant differences were analysed based on four technical replications (one‐way ANOVA; *P < 0.05). Bars indicate standard errors of the mean
Figure S20 The expression of two PR genes (PR1b and PR10a), two JA related genes (OsAOC and OsAOS2), one ET related gene (OsACS2), SA related genes (OsNPR1), and OsPAL in transgenic and WT plants after R. solani AG1‐IA infection at 72 hpi. The relative expression levels of OsAOC, OsAOS2, OsACS2, OsNPR1 and OsPAL are shown as the means of four lines. Statistically significant differences were analysed based on four technical replications (one‐way ANOVA; *P < 0.05). Bars indicate the standard errors of the mean
Figure S21 Changes in reactive oxygen species (ROS) levels of transgenic and WT
Figure S22 The interaction of 653 core candidate genes has been predicted
Figure S23 Concentrations of enzymes involve in the AsA‐GSH cycle in transgenic and WT; Teqing and Lemont) leaves at 72 hpi. Values of AsA, DHA, GSH, and GSSG activities represent the average of four lines. Data indicate mean ± standard error of mean (SEM) of four technical replicates. Statistically significant differences were analysed by one‐way ANOVA (*P < 0.05; **P < 0.01; ***P < 0.001)
Figure S24 The expression levels of OsAPX1, DHAR1, and GRX20 in transgenic and WT; Teqing and Lemont) leaves at 72 hpi. The rice UBQ gene was used as an internal control. Data are represented as average values with four biological replicates. Statistically significant differences were analysed by one‐way ANOVA (*P < 0.05; **P < 0.01; ***P < 0.001)
Figure S25 Changes in the disease resistance of CYP716A16‐OE transgenic rice plants. (a) Identification of OE lines of CYP716A16 by qRT–PCR. (b, c) Disease symptoms in CYP716A16‐OE plants and WT (Nipponbare) at 72 h after inoculation with AG1‐IA. Lesion represents the means of four lines. Statistically significant differences were analysed based on four technical replications (one‐way ANOVA; *P < 0.05; **P < 0.01; ***P < 0.001). Bars indicate standard errors of the mean
Figure S26 Changes in the disease resistance of OsSam12‐OE transgenic rice plants. (a) Identification of OE lines of OsSam12 by qRT–PCR. (b, c) Disease symptoms in OsSam12‐OE plants and WT (Nipponbare) at 72 h after inoculation with AG1‐IA. Lesion represents the means of four lines. Statistically significant differences were analysed based on four technical replications (one‐way ANOVA; *P < 0.05; **P < 0.01; ***P < 0.001). Bars indicate standard errors of the mean
Table S1 Names, origin, and population structure of 259 O. sativa accessions.
Table S2 Sequence information on the genomes of 259 rice accessions.
Table S3 Results of filtered SNP annotation.
Table S4 Number of SNPs in 259 rice accessions using the sequenced data mapped to the Nipponbare reference genome.
Table S5 GWAS of RSB resistant with 259 rice lines using the SNP‐set generated by mapping reads to the Nipponbare.
Table S6 The expression of SNP‐GWAS candidate genes (L: Lemont T: Teqing).
Table S7 The results from haplotype‐based GWAS of RSB resistant with 259 rice lines.
Table S8 The expression of Hap‐GWAS candidate genes (L: Lemons T:Teqing).
Table S9 The expression of DE candidate genes (L: Lemons T:Teqing).
Table S10 The different module gene set of WGCNA analyse (SNP‐GWAS candidate genes).
Table S11 The different module gene set of WGCNA analyse (HAP‐GWAS candidate gene).
Table S12 The different module gene set of WGCNA analyse (DEGs candidate genes).
Table S13 Final module candidate gene list and intersection gene (We chose the genes selected by the two methods as our final candidate gene set).
Table S14 Candidate genes expression and classification.
Table S15 Specific primer sequences used in our experiment.
Methods S1 Supplementary Methods.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (nos. 31400130, 31971867 and 32001490), the Sichuan Science and Technology Program (nos. 2019YFN0010 and 2020YFH0117), and the National 973 Project of China (no. 2014CB160304).
Wang, A., Shu, X., Jing, X., Jiao, C., Chen, L., Zhang, J., Ma, L., Jiang, Y., Yamamoto, N., Li, S., Deng, Q., Wang, S., Zhu, J., Liang, Y., Zou, T., Liu, H., Wang, L., Huang, Y., Li, P. and Zheng, A. (2021) Identification of rice (Oryza sativa L.) genes involved in sheath blight resistance via a genome‐wide association study. Plant Biotechnol. J., 10.1111/pbi.13569
Contributor Information
Ping Li, Email: liping6575@163.com.
Aiping Zheng, Email: apzh0602@gmail.com.
Data Availability
All the sequences have been deposited in the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under the accession PRJNA598020.
References
- Baggs, E., Dagdas, G. & Krasileva, K. V. (2017) NLR diversity, helpers and integrated domains: making sense of the NLR IDentity. Curr. Opin. Plant Biol. 38, 59–67. [DOI] [PubMed] [Google Scholar]
- Cesari, S., Thilliez, G., Ribot, C., Chalvon, V., Michel, C., Jauneau, A., Rivas, S.et al. (2013) The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR‐Pia and AVR1‐CO39 by direct binding. Plant Cell, 25, 1463–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W., Gao, Y.Q., Xie, W.B., Gong, L., Lu, K., Wang, W.S., Li, Y.et al. (2014) Genome‐wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat. Genet. 46, 714–721. [DOI] [PubMed] [Google Scholar]
- Chen, X.J., Chen, Y., Zhang, L.N., Xu, B., Zhang, J.H., Chen, Z.X., Tong, Y.H.et al. (2016) Overexpression of OsPGIP1 enhances rice resistance to sheath blight. Plant Dis. 100, 388–395. [DOI] [PubMed] [Google Scholar]
- Chen, Z.X., Feng, Z.M., Kang, H.X., Zhao, J.H., Chen, T.X., Li, Q.Q., Gong, H.B.et al. (2019) Identification of new resistance loci against sheath blight disease in rice through genome‐wide association study. Rice Sci. 26, 21–31. [Google Scholar]
- Conklin, P. L. (2001) Recent advances in the role and biosynthesis of ascorbic acid in plants. Plant Cell & Environment. 24, 383–394. [Google Scholar]
- Dilla‐Ermita, C.J., Tandayu, E., Juanillas, V.M., Detras, J., Lozada, D.N., Dwiyanti, M.S., Vera Cruz, C.et al. (2017) Genome‐wide association analysis tracks bacterial leaf blight resistance loci in rice diverse germplasm. Rice, 10, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam, K., Rao, P.B. and Chauhan, S.V.S. (2003) Efficacy of some botanicals of the family compositae against Rhizoctonia solani Kuhn. J. Mycol. Plant Pathol. 33, 230–235. [Google Scholar]
- Huang, X.H., Wei, X.H., Sang, T., Zhao, Q., Feng, Q., Zhao, Y., Li, C.Y.et al. (2010) Genome‐wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 42, 961–967. [DOI] [PubMed] [Google Scholar]
- Iwai, T., Miyasaka, A., Seo, S. and Ohashi, Y. (2006) Contribution of ethylene biosynthesis for resistance to blast fungus infection in young rice plants. Plant Physiol. 142, 1202–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, Y., McAdams, S.A., Bryan, G.T., Hershey, H.P. and Valent, B. (2000) Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, H.W., Panigrahi, G.K., Jung, G.Y., Lee, Y.J., Shin, K.H., Sahoo, A., Choi, E.S.et al. (2020) Pathogen‐associated molecular pattern‐triggered immunity involves proteolytic degradation of core nonsense‐mediated mRNA decay factors during the early defense response. Plant Cell, 32, 1081–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmakar, S., Molla, K.A., Chanda, P.K., Sarkar, S.N., Datta, S.K. and Datta, K. (2016) Green tissue‐specific co‐expression of chitinase and oxalate oxidase 4 genes in rice for enhanced resistance against sheath blight. Planta, 243, 115–130. [DOI] [PubMed] [Google Scholar]
- Kawano, Y., Akamatsu, A., Hayashi, K., Housen, Y., Okuda, J., Yao, A., Nakashima, A.et al. (2010) Activation of a Rac GTPase by the NLR family disease resistance protein Pit plays a critical role in rice innate immunity. Cell Host Microbe, 7, 362–375. [DOI] [PubMed] [Google Scholar]
- Kim, Y.S., Kim, I.S., Bae, M.J., Choe, Y.H., Kim, Y.H., Park, H.M., Kang, H.G.et al. (2013) Homologous expression of cytosolic dehydroascorbate reductase increases grain yield and biomass under paddy field conditions in transgenic rice (Oryza sativa L. japonica). Planta, 237, 1613–1625. [DOI] [PubMed] [Google Scholar]
- Lavale, S.A., Prashanthi, S.K. and Fathy, K. (2018) Mapping association of molecular markers and sheath blight (Rhizoctonia solani) disease resistance and identification of novel resistance sources and loci in rice. Euphytica, 214, 78. [Google Scholar]
- Lee, F.N. and Rush, M.C. (1983) Rice sheath blight: A major rice disease. Plant Dis. 67, 829–832. [Google Scholar]
- Li, N., Lin, B., Wang, H., Li, X.M., Yang, F.F., Ding, X.H., Yang, J.B.et al. (2019) Natural variation in ZmFBL41 confers banded leaf and sheath blight resistance in maize. Nat. Genet. 51, 1540–1548. [DOI] [PubMed] [Google Scholar]
- Li, R., Afsheen, S., Xin, Z.J., Han, X. and Lou, Y.G. (2013) OsNPR1 negatively regulates herbivore‐induced JA and ethylene signaling and plant resistance to a chewing herbivore in rice. Physiol. Plant. 147, 340–351. [DOI] [PubMed] [Google Scholar]
- Liu, G., Jia, Y., Correa‐Victoria, F., Prado, G., Yeater, K. and McClung, A. (2009) Mapping quantitative trait loci responsible for resistance to sheath blight in rice. Phytopathology, 99, 1078–1084. [DOI] [PubMed] [Google Scholar]
- Liu, S., Wang, Z.B., Chen, D., Zhang, B.W., Tian, R.R., Wu, J., Zhang, Y.et al. (2017) Annotation and cluster analysis of spatiotemporal‐ and sex‐related lncRNA expression in rhesus macaque brain. Genome Res. 27, 1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W., Liu, J., Triplett, L., Leach, J.E. and Wang, G.L. (2014) Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu. Rev. Phytopathol. 52, 1–29. [DOI] [PubMed] [Google Scholar]
- van Loon, L.C., Rep, M. and Pieterse, C.M.J. (2006) Significance of inducible defense‐related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. [DOI] [PubMed] [Google Scholar]
- Ma, Z.Y., He, S.P., Wang, X.F., Sun, J.L., Zhang, Y., Zhang, G.Y., Wu, L.Q.et al. (2018) Resequencing a core collection of upland cotton identifies genomic variation and loci influencing fiber quality and yield. Nat. Genet. 50, 803–813. [DOI] [PubMed] [Google Scholar]
- Marone, D., Russo, M.A., Laidò, G., De Leonardis, A.M. and Mastrangelo, A.M. (2013) Plant nucleotide binding site‐leucine‐rich repeat (NBS‐LRR) genes: active guardians in host defense responses. Int. J. Mol. Sci. 14, 7302–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale, L., Tan, X.P., Koehl, P. and Michelmore, R.W. (2006) Plant NBS‐LRR proteins: adaptable guards. Genome Biol. 7, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei, C.S., Qi, M., Sheng, G.Y. and Yang, Y.N. (2006) Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol. Plant Microbe Interact. 19, 1127–1137. [DOI] [PubMed] [Google Scholar]
- Michael, R., Ken, H., Takafumi, S., Kazunori, O., Sugihiro, A., Susumu, M., Yoko, N.et al. (2013) Identification of rice allene oxide cyclase mutants and the function of jasmonate for defence against Magnaporthe oryzae . Plant J. 74, 226–238. [DOI] [PubMed] [Google Scholar]
- Miyashita, Y., Takasugi, T. and Ito, Y. (2010) Identification and expression analysis of PIN genes in rice. Plant Sci. 178, 424–428. [Google Scholar]
- Molla, K.A., Karmakar, S., Molla, J., Bajaj, P., Varshney, R.K., Datta, S.K. and Datta, K. (2020) Understanding sheath blight resistance in rice: the road behind and the road ahead. Plant Biotechnol. J. 18, 895–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oreiro, E.G., Grimares, E.K.S., Grande, G.A., Quibod, L.L., Roman Reyna, V. and Oliva, R.F. (2019) Genome‐wide associations and transcriptional profiling reveal ROS regulation as one underlying mechanism of sheath blight resistance in rice. Mol. Plant Microbe Interact. 33, 212–222. [DOI] [PubMed] [Google Scholar]
- Peng, X.X., Hu, Y.J., Tang, X.K., Zhou, P.L., Deng, X.B., Wang, H.H. and Guo, Z.J. (2012) Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta, 236, 1485–1498. [DOI] [PubMed] [Google Scholar]
- Peng, X.X., Wang, H.H., Jang, J.C., Xiao, T., He, H.H., Jiang, D. and Tang, X.K. (2016) OsWRKY80‐OsWRKY4 module as a positive regulatory circuit in rice resistance against Rhizoctonia solani . Rice, 9, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinson, S.R.M., Capdevielle, F.M. and Oard, J.H. (2005) Confirming QTLs and finding additional loci conditioning sheath blight resistance in rice using recombinant inbred lines. Crop Sci. 45, 503–510. [Google Scholar]
- Rohini, G., Shalu, J., Akhilesh, K.T. and Mukesh, J. (2010) Genome‐wide survey and expression analysis suggest diverse roles of glutaredoxin gene family members during development and response to various stimuli in rice. DNA Res. 17, 353–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, F., Golstein, C., Ade, J., Stoutemyer, M., Dixon, J.E. and Innes, R.W. (2003) Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science, 301, 1230–1233. [DOI] [PubMed] [Google Scholar]
- Shen, Q.H., Saijo, Y., Mauch, S., Biskup, C., Bieri, S., Keller, B., Seki, H.et al. (2007) Nuclear activity of MLA immune receptors links isolate‐specific and basal disease‐resistance responses. Science, 315, 1098–1103. [DOI] [PubMed] [Google Scholar]
- Srinivasachary, W.L. and Savary, S. (2011) Resistance to rice sheath blight (Rhizoctonia solani Kuhn) [(Teleomorph: Thanatephorus cucumeris (A.B. Frank) Donk.] disease: current status and perspectives. Euphytica, 178, 1–22. [Google Scholar]
- Ströher, E. and Millar, A.H. (2012) The biological roles of glutaredoxins. Biochem J. 446, 333–348. [DOI] [PubMed] [Google Scholar]
- Sun, Q., Li, T.Y., Li, D.D., Wang, Z.Y., Li, S., Li, D.P., Han, X.et al. (2019) Overexpression of loose plant architecture 1 increases planting density and resistance to sheath blight disease via activation of PIN‐FORMED 1a in rice. Plant Biotechnol. J. 17, 855–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X., Wang, P., Jia, X., Huo, L., Che, R. and Ma, F. (2018) Improvement of drought tolerance by overexpressing MdATG18a is mediated by modified antioxidant system and activated autophagy in transgenic apple. Plant Biotechnol. J. 16, 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, H., Chen, Z., Du, Z., Liu, Y. and Klessig, D.F. (1997) Development of necrosis and activation of disease resistance in transgenic tobacco plants with severely reduced catalase levels. Plant J. 11, 993–1005. [DOI] [PubMed] [Google Scholar]
- Tonnessen, B.W., Manosalva, P., Lang, J.M., Baraoidan, M., Bordeos, A., Mauleon, R., Oard, J.et al. (2015) Rice phenylalanine ammonia‐lyase gene OsPAL4 is associated with broad spectrum disease resistance. Plant Mol. Biol. 87, 273–286. [DOI] [PubMed] [Google Scholar]
- Van‐Nas, A., Thakurta, D.G., Wang, S.S., Yehya, N., Horvath, S., Zhang, B., Ingram‐Drake, L.et al. (2009) Elucidating the role of gonadal hormones in sexually dimorphic gene co‐expression networks. Endocrinology, 150, 1235–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D.K., Liu, H.Q., Li, S.J., Zhai, G.W., Shao, J.F. and Tao, X.Z. (2015) Characterization and molecular cloning of a serine hydroxyl methyl transferase 1 (OsSHM1) in rice. J. Integr. Plant Biol. 57, 745–756. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Pinson, S.R.M., Fjellstrom, R.G. and Tabien, R.E. (2012) Phenotypic gain from introgression of two QTL, qSB9‐2 and qSB12‐1 for rice sheath blight resistance. Mol. Breed. 30, 293–303. [Google Scholar]
- Wu, C. H., Krasileva, K. V., Banfield, M. J., Terauchi, R. & Kamoun, S. (2015) The "sensor domains" of plant NLR proteins: more than decoys. Front Plant Sci. 6, 134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, X., Cao, Z.X., Zhang, X.T., Wang, Y. and Zuo, S.M. (2016) Overexpression of OsOSM1 enhances resistance to rice sheath blight. Plant Dis. 100, 1634–1642. [DOI] [PubMed] [Google Scholar]
- Yuan, B., Zhai, C., Wang, W.J., Zeng, X.S., Xu, X.K., Hu, H.Q., Lin, F.et al. (2011) The Pik‐p resistance to Magnaporthe oryzae in rice is mediated by a pair of closely linked CC‐NBS‐LRR genes. Theor. Appl. Genet. 122, 1017–1028. [DOI] [PubMed] [Google Scholar]
- Zhang, F., Wu, Z.C., Wang, M.M., Zhang, F., Dingkuhn, M., Xu, J.L., Zhou, Y.L.et al. (2017a) Genome‐wide association analysis identifies resistance loci for bacterial blight in a diverse collection of indica rice germplasm. PLoS One, 12, e0174598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J.F., Chen, L., Fu, C.L., Wang, L.X., Liu, H.N., Cheng, Y.Z., Li, S.C.et al. (2017b) Comparative transcriptome analyses of gene expression changes triggered by Rhizoctonia solani AG1 IA infection in resistant and susceptible rice varieties. Front. Plant Sci. 8, 1422–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.C. and Gassmann, W. (2003) RPS4‐Mediated disease resistance requires the combined presence of RPS4 transcripts with full‐length and truncated open reading frames. Plant Cell, 15, 2333–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z.Z., Pang, Z.Q., Zhao, S.L., Zhang, L.L., Lv, Q.M., Yin, D.D., Li, D.Y.et al. (2019) Importance of OsRac1 and RAI1 in signalling of NLR protein‐mediated resistance to rice blast disease. New Phytol. 223, 828–838. [DOI] [PubMed] [Google Scholar]
- Zou, J.H., Pan, X.B., Chen, Z.X., Xu, J.Y., Lu, J.F., Zhai, W.X. and Zhu, L.H. (2000) Mapping quantitative trait loci controlling sheath blight resistance in two rice cultivars (Oryza sativa L.). Theor. Appl. Genet. 101, 569–573. [Google Scholar]
- Zuo, S.M., Yin, Y.J., Zhang, L., Zhang, Y.F., Chen, Z.X. and Pan, X.B. (2007) Breeding value and further mapping of a QTL qSB‐11 conferring the rice sheath blight resistance. Chinese J. Rice Sci. 21, 136–142. [Google Scholar]
- Zuo, S.M., Yin, Y.J., Pan, C.H., Chen, Z.X., Zhang, Y.F., Gu, S.L., Zhu, L.H.et al. (2013) Fine mapping of qSB‐11LE, the QTL that confers partial resistance on rice sheath blight. Theor. Appl. Genet. 126, 1257–1272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 The geographic distribution of 259 rice accessions. Each dot of a given colour on the world map represents the geographic distribution of the corresponding rice accession
Figure S2 Distribution of single nucleotide polymorphisms (SNPs) and nucleotide diversity across the rice Nipponbare genome in the rice association panel
Figure S3 The frequency distribution of incidence rate of 259 rice lines after inoculation with Rhizoctonia solani AG1‐IA
Figure S4 Histograms and box plots showing the resistance phenotypic data of 259 rice lines at three grown stages to rice sheath blight (RSB)
Figure S5 Manhattan plots and QQ plots resulting from the SNP‐based GWAS for RSB resistance
Figure S6 Several known rice sheath blast resistant genes were identified by SNP‐based and haplotype‐based GWAS
Figure S7 Manhattan plots resulting from the haplotype‐based GWAS for RSB resistance
Figure S8 Chromosomal positions of loci for RSB resistance identified via SNP‐based and haplotype‐based GWAS. (a) Seedling stage. (b) Tillering stage. (c) Booting stage. The mapped loci in this study are shown on the right, and previously mapped QTLs are shown in pink region
Figure S9 WGCNA of genes in SNP‐GWAS, haplotype‐based GWAS and DEGs three datasets
Figure S10 The expression profiles and pathway analysis of 653 core candidate RSB resistant genes in this study
Figure S11 Manhattan plots for RSB resistant on chromosome 11
Figure S12 PCR amplification results of OsRSR1 in resistant and susceptible rice line. M: DNA marker; 1‐14 represent ID of rice lines 267, 136, 139, 142, 158, 178, 14, 283, 202, 307, 197, 199, 208, and 323, respectively
Figure S13 Phylogenetic and structural analysis of two candidate resistant genes. (a) Phylogenetic analysis of the deduced amino acid sequence of OsRSR1 and OsRLCK5 compared with Arabidopsis thaliana and rice sequences. (b) Schematic diagram of the OsRSR1 and OsRLCK5 proteins
Figure S14 Changes in the disease resistance of OsRSR1‐RNAi and OsRSR1‐overexpressing (OE) transgenic rice plants. Leaf lesion represents the means of four lines. Statistically significant differences were analysed based on four technical replications (one‐way ANOVA; ***P < 0.001). Bars indicate standard errors of the mean
Figure S15 The plant height, number of tillers, setting percentage, and 1,000‐grain weight of OE transgenic plant and wild type (WT) were observed
Figure S16 (a) Expression analysis of OsRLCK5 and OsRSR1 in WT and OE transgenic T2 rice plants by qRT–PCR. (b) Changes in the disease resistance of OsRSR1 and OsRLCK5 T2 transgenic lines. Leaf lesion represents the means of five lines. Statistically significant differences were analysed based on four technical replications (one‐way ANOVA; **P < 0.01, ***P < 0.001). Bars indicate standard errors of the mean
Figure S17 Manhattan plots for RSB resistant on chromosome 01
Figure S18 Alignment of the conserved serine‐threonine domain of OsRLCK5, its homologs in Arabidopsis thaliana, and other rice serine‐threonine kinases. Underlined regions represent the conserved subdomains of serine/threonine protein kinases; the residues marked with asterisks are the three phosphorylated catalytic sites
Figure S19 Changes in the disease resistance of OsRLCK5‐RNAi and OsRLCK5‐OE transgenic rice plants. (a) Disease symptoms in TeQing, Lemont, OsRLCK5‐RNAi and OsRLCK5‐OE plants at 72 h after inoculation with AG1 IA. Leaf lesion represents the means of four lines. (b) Disease severity was evaluated as relative lesion area at 72 hour postinoculation (hpi). Statistically significant differences were analysed based on four technical replications (one‐way ANOVA; *P < 0.05). Bars indicate standard errors of the mean
Figure S20 The expression of two PR genes (PR1b and PR10a), two JA related genes (OsAOC and OsAOS2), one ET related gene (OsACS2), SA related genes (OsNPR1), and OsPAL in transgenic and WT plants after R. solani AG1‐IA infection at 72 hpi. The relative expression levels of OsAOC, OsAOS2, OsACS2, OsNPR1 and OsPAL are shown as the means of four lines. Statistically significant differences were analysed based on four technical replications (one‐way ANOVA; *P < 0.05). Bars indicate the standard errors of the mean
Figure S21 Changes in reactive oxygen species (ROS) levels of transgenic and WT
Figure S22 The interaction of 653 core candidate genes has been predicted
Figure S23 Concentrations of enzymes involve in the AsA‐GSH cycle in transgenic and WT; Teqing and Lemont) leaves at 72 hpi. Values of AsA, DHA, GSH, and GSSG activities represent the average of four lines. Data indicate mean ± standard error of mean (SEM) of four technical replicates. Statistically significant differences were analysed by one‐way ANOVA (*P < 0.05; **P < 0.01; ***P < 0.001)
Figure S24 The expression levels of OsAPX1, DHAR1, and GRX20 in transgenic and WT; Teqing and Lemont) leaves at 72 hpi. The rice UBQ gene was used as an internal control. Data are represented as average values with four biological replicates. Statistically significant differences were analysed by one‐way ANOVA (*P < 0.05; **P < 0.01; ***P < 0.001)
Figure S25 Changes in the disease resistance of CYP716A16‐OE transgenic rice plants. (a) Identification of OE lines of CYP716A16 by qRT–PCR. (b, c) Disease symptoms in CYP716A16‐OE plants and WT (Nipponbare) at 72 h after inoculation with AG1‐IA. Lesion represents the means of four lines. Statistically significant differences were analysed based on four technical replications (one‐way ANOVA; *P < 0.05; **P < 0.01; ***P < 0.001). Bars indicate standard errors of the mean
Figure S26 Changes in the disease resistance of OsSam12‐OE transgenic rice plants. (a) Identification of OE lines of OsSam12 by qRT–PCR. (b, c) Disease symptoms in OsSam12‐OE plants and WT (Nipponbare) at 72 h after inoculation with AG1‐IA. Lesion represents the means of four lines. Statistically significant differences were analysed based on four technical replications (one‐way ANOVA; *P < 0.05; **P < 0.01; ***P < 0.001). Bars indicate standard errors of the mean
Table S1 Names, origin, and population structure of 259 O. sativa accessions.
Table S2 Sequence information on the genomes of 259 rice accessions.
Table S3 Results of filtered SNP annotation.
Table S4 Number of SNPs in 259 rice accessions using the sequenced data mapped to the Nipponbare reference genome.
Table S5 GWAS of RSB resistant with 259 rice lines using the SNP‐set generated by mapping reads to the Nipponbare.
Table S6 The expression of SNP‐GWAS candidate genes (L: Lemont T: Teqing).
Table S7 The results from haplotype‐based GWAS of RSB resistant with 259 rice lines.
Table S8 The expression of Hap‐GWAS candidate genes (L: Lemons T:Teqing).
Table S9 The expression of DE candidate genes (L: Lemons T:Teqing).
Table S10 The different module gene set of WGCNA analyse (SNP‐GWAS candidate genes).
Table S11 The different module gene set of WGCNA analyse (HAP‐GWAS candidate gene).
Table S12 The different module gene set of WGCNA analyse (DEGs candidate genes).
Table S13 Final module candidate gene list and intersection gene (We chose the genes selected by the two methods as our final candidate gene set).
Table S14 Candidate genes expression and classification.
Table S15 Specific primer sequences used in our experiment.
Methods S1 Supplementary Methods.
Data Availability Statement
All the sequences have been deposited in the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under the accession PRJNA598020.


