Figure 1.
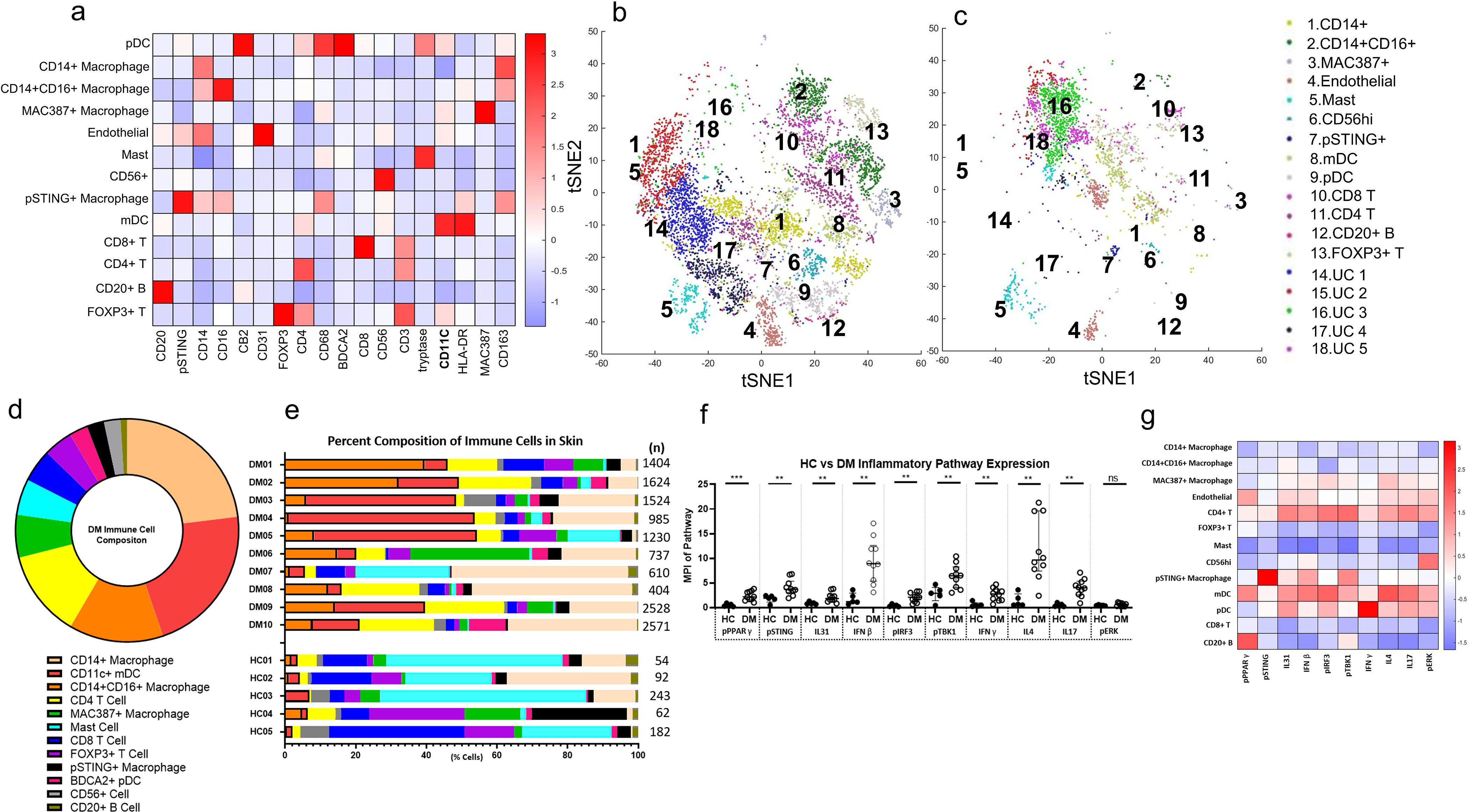
Unbiased Clustering of Immune Cells in DM Skin Lesions using PhenoGraph a) Heatmap of Different Cell Type Markers expressed the 13 cell populations identified: CD14+ macrophages, CD14+CD16+ macrophages, MAC387+ macrophages, CD31+ endothelial cells, CD4+ T cells, FOXP3+ Tregs, tryptase+ mast cells, CD56+ cells, pSTING+ macrophages, CD11c+ mDCs, BDCA2+ pDCs, and CD8+ T cells, and CD20+ B cells. b) tSNE dimensional reduction plot of the different cell clusters identified in lesional DM skin. c) tSNE dimensional reduction plot of the corresponding cell clusters in HC skin. Clusters 14–18 represent unidentified clusters (UC). d) Average percent composition of immune cells identified in DM skin from DM1-DM10 (%): CD14+ macrophage (23.0), CD11C+ mDC (21.7), CD14+CD16+ macrophage (13.7), CD4 T Cell (12.6), MAC387+ macrophage (6.2), mast (5.2), CD8 T Cell (4.8), FOXP3+ T Cell (4.2), pSTING+ macrophage (2.5), BDCA2+ pDC (2.4), CD56+ cells (2.4), and CD20+ B Cell (1.0) Data are represented as mean ± SEM. e) The percent composition of immune cells identified in each DM patient and HC. f) Expression of pPPARγ, pSTING, pIRF3, pTBK1, IFNβ, IFNγ, IL4, IL17, and IL31 is increased in DM skin compared to HC skin. There is no significant difference between DM and HC skin pERK expression. g) Heatmap displaying the relative mean expression of each inflammatory pathway in each cell cluster. Data are represented as median ± IQR. * p<0.05, ** p<0.01, *** p<0.001
