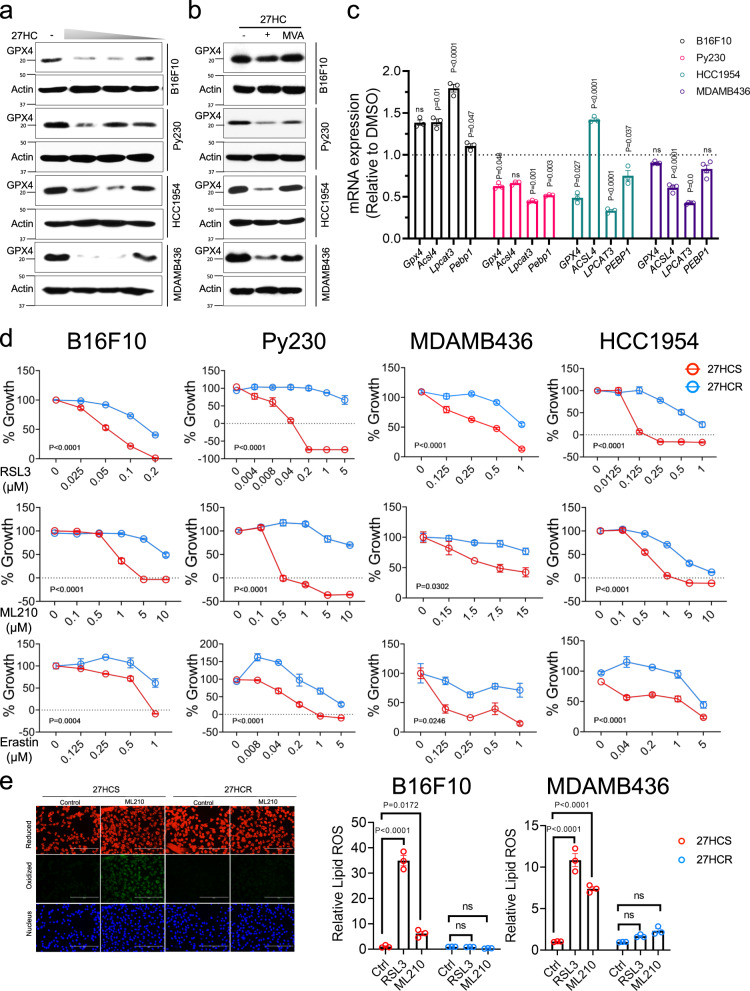
Fig. 4. 27HCR cells are resistant to ferroptosis.
a B16F10, Py230, HCC1954, and MDAMB436 cells were treated with various doses of 27HC for 48 hr. Cell lysates were harvested, and western immunoblots were used to analyze GPX4 expression levels. β-Actin was used as a loading control. Representative results from two independent experiments are shown. b mevalonate (MVA, 500 µM) supplementation reversed 27HC-dependent inhibition of GPX4 protein expression in B16F10, Py230, MDAMB436, and HCC1954 cells. Representative results from three independent experiments are shown. c qRT-PCR analysis of the expression of genes involved in lipid peroxidation (GPX4, Acsl4, Lpcat3, and Pebp1) in 27HCS and 27HCR cells. Data plotted as mean ± SEM (n = 3 wells of cells for B16F10, Py230, and HCC1954; n = 4 wells of cells for MDAMB436). P values were calculated using two-sided unpaired Student’s t test. d 27HCS and 27HCR derivatives of B16F10, Py230, HCC1954, and MDAMB436 cells were treated with inducers of ferroptosis, including GPX4 inhibitors, RSL3 (upper), and ML210 (middle), and the xc- inhibitor, erastin (lower) and 0.1% DMSO control. Morphology of ferroptotic cell death such as cells rounding up and plasma membrane rupture were observed in treated cells. Cell growth was assessed after 48–72 h by measuring DNA content using Hoechst 33258. Relative cell growth (% of Growth) was then calculated by normalizing each treatment condition to control-treated wells. Data plotted as mean ± SEM as representative results from four independent experiments (n = 5 wells of cells for B16F10, MDAMB436, and HCC1954; n = 3 wells of cells for Py230). P values were calculated using two-way ANOVA. e RSL3 (1 µM) and ML210 (5 µM) induced lipid peroxidation in 27HCS and 27HCR-detivatives of B16F10 and -MDAMB436 cells as measured by BODIPY-C11 staining. Fluorescent images of BODIPY-C11 stained B16F10 cells treated with ML210 for 4 h are shown in the left panels. Data plotted as mean ± SEM; n = 3 wells of cells. P values were calculated using one-way ANOVA. Figure 4e results were repeated in Py230 cells. Unprocessed immunoblots and numerical source data are reported in the Source Data File.