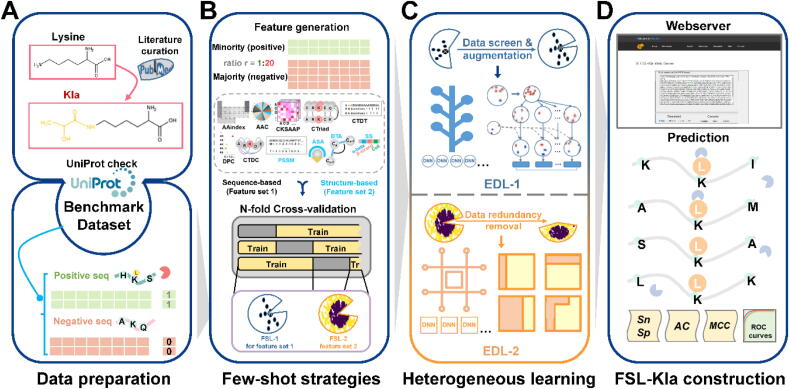
Fig. 1.
The protocol of this study. A. From the latest literature, we obtained and collected 343 unique Kla sites as the dataset. All the entries must have a PMID and specify a clear position for Kla. Then we corrected the above dataset by UniProt and formed the benchmark dataset. All the entries are labeled and the non-Kla sites in the same protein or peptide are regarded as negative samples. B. The feature encoding schemes for both feature set 1 and set 2 with their corresponding imbalance strategy. The generated feature encodings including AAC, CTriad, AAindex, DPC, CTDT, CTDC, CKSAPP are grouped in feature set 1 while other four encodings ASA, BTA, PSSM and SS are grouped in feature set 2. Stratified cross-validations with few-shot strategies were conducted. FSL-1 was applied in feature set 1 while only FSL-2 was applied in feature set 2 because of some principles of few-shot strategies. C. The diagram of EDL-1 for feature set 1 and EDL-2 for feature set 2. The upper cell shows the strategic combination for both major and minor class and then a vote determines the combinatory results. The lower cell shows the process of ensemble. Samples that are wrongly classified will attain higher weight values in the next iteration. The final box is an optimized box based on many base classifiers. An adaptive decision boundary was also shown in the final box. D. The construction of FSL-Kla webserver and some evaluation metrics of Kla sites prediction.