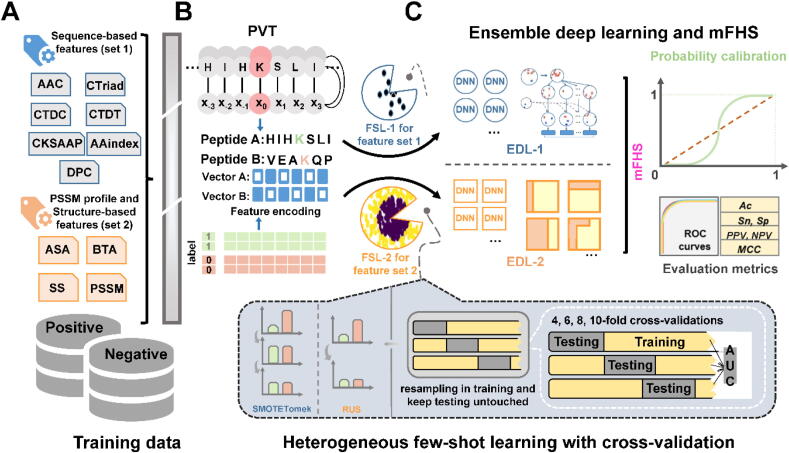
Fig. 2.
Architecture and implemental steps of the ensemble method with imbalance strategy. A. Data collection and feature encoding schemes for both amino acid composition-based features as well as PSSM profile and structure-based features. B. Generating encodings for labeled proteins and peptides: A procedure for peptides to vectors transformation (PVT). C. Implementation of FSL-1 with EDL-1 for feature set 1 while FSL-2 with EDL-2 for feature set 2. Probability calibration performs an important role in correcting prediction after primary ensemble. Base learners’ outputs after probability calibration are stacked as information for the input of mFHS. The 4, 6, 8 and 10-fold stratified cross-validations were executed to evaluate the performance. Conducting few-shot strategies in training data and keeping testing data untouched provides a reasonable evaluation approach from the data aspect. ROC curves with some evaluation metrics including Sn, Sp, Ac, PPV, NPV and MCC are utilized.