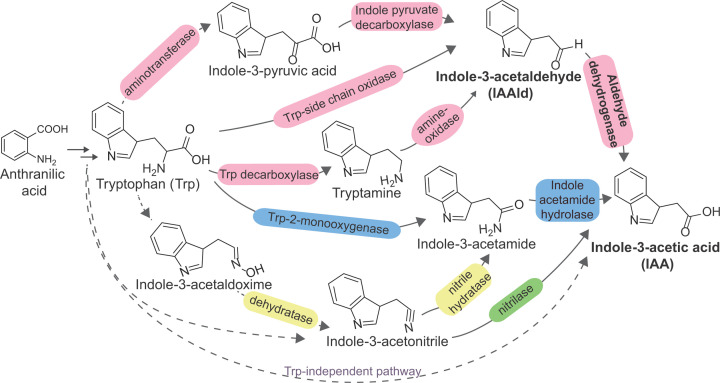
Figure 1. Bacterial biosynthesis of auxin.
Certain plant-associated bacteria can produce auxin indole-3-acetic acid (IAA) from tryptophan (Trp) as a precursor using one or more than one of the routes shown here. Among the known Trp-dependent pathways, three routes involving (i) aminotransferase and indole-3-pyruvate decarboxylase, (ii) Trp-side chain oxidase, and (iii) Trp deacarboxylase and an amine-oxidase converge at indole-3-acetaldehyde (IAAld) which is then oxidized by aldehyde dehydrogenase (ALDH) to yield IAA (enzymes shown in pink). Another prominent route involves the intermediate indole-3-acetamide which can be synthesized directly from tryptophan by tryptophan 2-monooxygenase (iaaM), and then converted into IAA via the enzyme indole acetamide hydrolase (iaaH) (enzymes shown in blue). Alternately, indole-3-acetamide is synthesized via a two-step reaction involving a predicted dehydratase and nitrile hydratase (enzymes shown in yellow) followed by conversion into IAA as mentioned previously. Finally, some bacteria appear to use the lesser studied Trp-independent routes, though the precursors appear to be derived from Trp biosynthesis intermediates [16,17]. For example, indole-3-acetonitrile, derived from a precursor of Trp yield IAA in a single-step reaction catalyzed by the nitrilase enzyme (enzyme shown in green). The reactions for which genetic and biochemical evidence of the enzymes involved are yet to discovered are shown with dashed arrows. Pseudomonas syringae DC3000 genome possesses genes for amine oxidase, nitrilase, indole acetamide hydrolase, ALDH, and a putative monooxygenase [19]. The enzymes shown with dashed arrows have some biochemical evidence however, the encoding genes are yet unknown. Figure adapted from Spaepen and Vanderleyden (2011) [15] and Duca et al. (2014) [18].