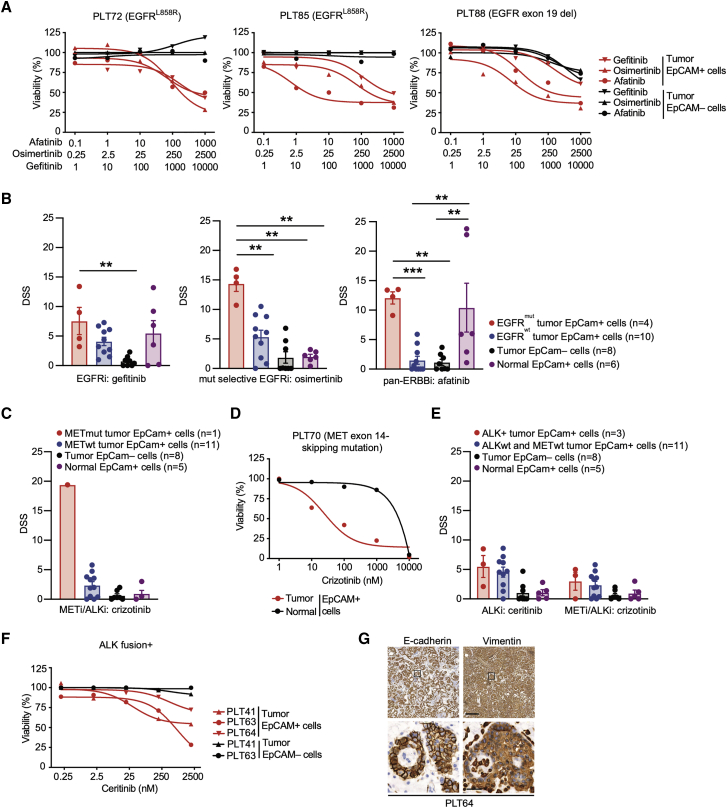
Figure 4.
NSCLC FUTCs predict genotype-matched therapeutic responses
(A) Dose-response curves of gefitinib, osimertinib, and AF in patient-matched tumor-derived EpCAM+ and EpCAM– cells.
(B) DSS for gefitinib, osimertinib, and AF compared between EGFR mutant EpCAM+, EGFR wild-type EpCAM+, tumor EpCAM–, and normal EpCAM+ cells. Each dot represents an independent sample.
(C and D) Bar graph representing the DSS of crizotinib (C) and (D) ceritinib and crizotinib across all of the patient samples.
(E) Dose-response curves of crizotinib in patient-matched tumor-derived EpCAM+ or normal tissue-derived EpCAM+ cells.
(F) Dose-response curves of ceritinib in patient-matched tumor-derived EpCAM+ and EpCAM– cells.
(G) Representative IHC images of E-cadherin and vimentin staining in patient (PLT64)-derived EML4-ALK+ lung tumor tissues. Scale bars correspond to 200 or 20 μm for low or high magnifications, respectively. Boxes indicate areas shown in the higher magnification in a lower row.
Error bars represent ±SEMs. Two-tailed unpaired Student’s t test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.