Figure 4.
Autophagic clearance and mitochondrial quality and functions are impaired in human lipin1-deficient myoblasts
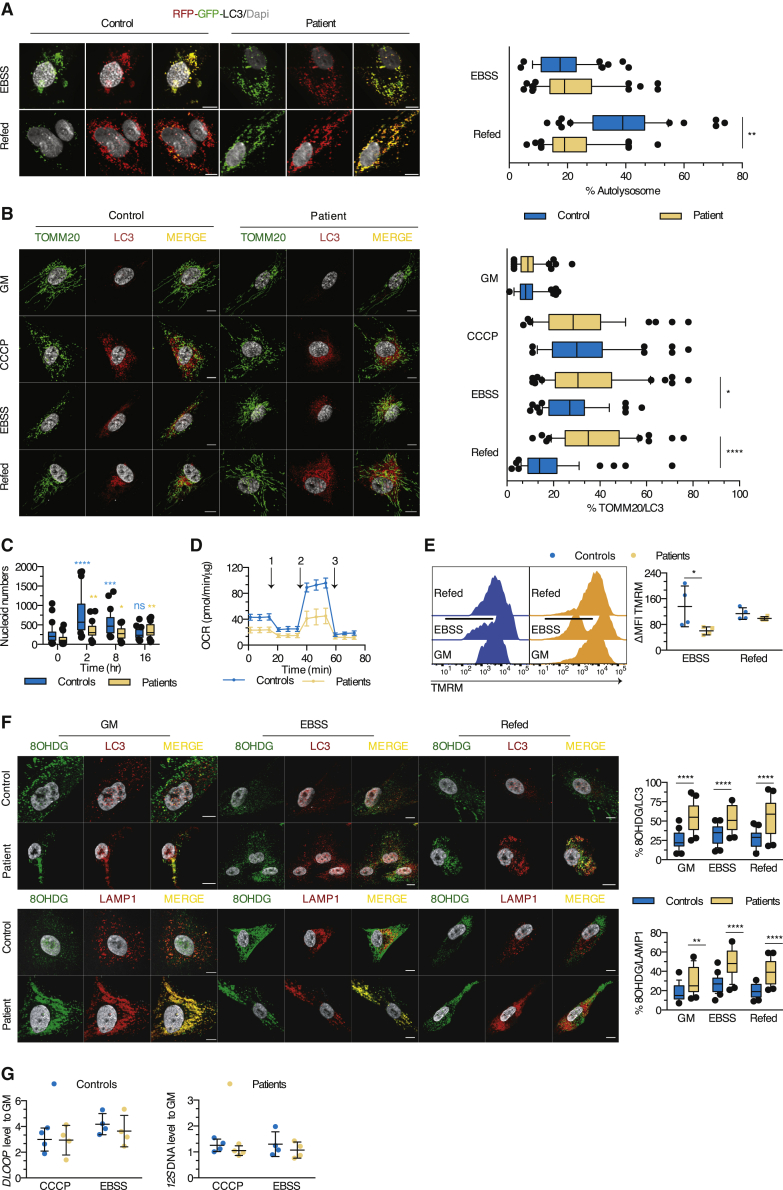
(A) Autolysosome formation (red puncta) in myoblasts transfected with the RFP-GFP-LC3 construct upon exposure to EBSS and refed or not with GM. Box and whisker plots (50 images/condition) represent the percentage of autolysosomes (mean effect of interaction F(1,196) = 44.4, p < 0.0001, of stimulus F(1,196) = 47.63, p < 0.0001, of subjects F(1,196) = 17.78, p < 0.0001).
(B) Myoblasts were cultured in GM or EBSS and refed or not with GM (starvation-induced mitophagy) or exposed to CCCP before immunostaining for TOMM20 and LC3. Box and whisker plots (50 images/condition) depict the percentage of proximity of TOMM20 with LC3 (mean effect of interaction F(3,392) = 11.09, p < 0.0001, of stimulus F(3,392) = 62.9, p < 0.0001, of subjects F(1,392) = 20.70, p < 0.0001).
(C) Box and whisker plots (30 images/condition) represent the number of mitochondrial nucleoids per myoblast after exposure to CCCP (mean effect of interaction F(3,116) = 9.554, p < 0.0001, of time F(3,116) = 19.99, p < 0.0001, of subjects F(1;116) = 26.22, p < 0.0001).
(D) Real-time analysis of oxygen consumption rate (OCR) in myoblasts under basal respiration and after addition of (1) oligomycin, (2) FCCP, or (3) antimycin A. Dot plot (mean of 4–6 technical replicates/dot) represents the means ± SDs of the OCR in myoblasts from 5 controls and 4 patients.
(E) Evaluation of the mitochondrial membrane potential by flow cytometry. Dot plot (mean of 2 technical replicates/dot) depicts the means ± SDs of the MFI of TMRM, expressed as a percentage of the MFI for GM condition (mean effect of interaction F(1,12) = 3.340, p = 0.0926, of stimulus F(1,12) = 7.313, p = 0.0192, of subjects F(1,12) = 0.2404, p = 0.6327).
(F) Distribution of oxidized DNA within the LC3 and LAMP1 structures of myoblasts cultured as in (B). Box and whisker plots (25 images/condition) show the percentage of proximity of 8OHDG with LC3 (mean effect of interaction F(2,144) = 1.504, p = 0.2258, of stimulus F(2,144) = 0.2997, p = 0.7415, of subjects F(1,144) = 100.8, p < 0.0001) or LAMP1 (mean effect interaction F(2,144) = 3.130, p = 0.0467, of stimulus F(2,144) = 18.41, p < 0.0001, of subjects F(1,144) = 96.93, p < 0.0001).
(G) Quantification of 12S mtDNA levels qPCR in cytosolic fractions from myoblasts of 4 patients and controls exposed to CCCP or EBSS (mean effect of interaction F(1,12) = 0.004867, p = 0.9455, of stimulus F(1,12) = 1.798, p = 0.2047, of subjects F(1,12) = 0.03755, p = 0.8496) and of the mtDNA motif DLOOP (mean effect of interaction F(1,12) = 0.2168, p = 0.6498, of stimulus F(1,12) = 0.3160, p = 0.5844, of subjects F(1,12) = 3.371, p = 0.0913). Dot plots (mean of 3 technical replicates/dot) depict the means ± SDs of the ratio of cycle threshold (CT) values of the given cytosolic fraction normalized to unfractioned cells.
Scale bars, 10 μm (A and B). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001: adjusted p values after between-subjects (A, B, and E–G) or within-subjects (C, as compared to H0 for each family) 2-way ANOVA and post hoc Sidak’s correction for multiple comparisons. Images and plots show typical staining and quantification for 1 of 3 patients and controls (A, B, C, and F). Results are representative from 1 of 2 (G), 3 (A–D and F), and 4 (E) independent experiments.
See also Figures S3 and S4.