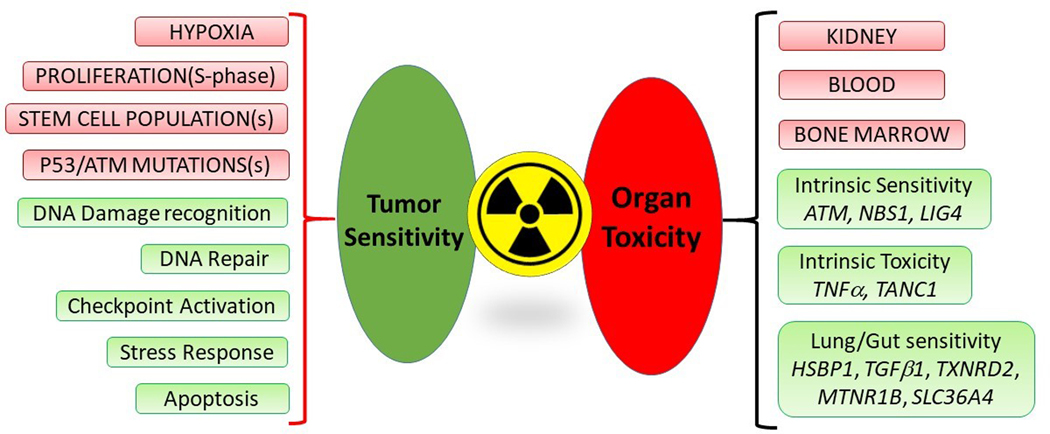
Figure 1. Radiation sensitivity and toxicity: pathobiological features, pathways and candidate genes.
Tumor features relevant to radiation sensitivity (pink – top left): include proliferation, hypoxia, “stemness” and mutations particularly in TP53 and the ATM genes. Molecular profiling studies have identified pathways common to radiation responsiveness (green – bottom left). These include DNA damage recognition and repair; senescence induction and apoptosis.
Radiation toxicity affects the kidneys, individual blood cell populations e.g., platelets, white cells and the bone marrow (pink – top right). Candidate factors related to toxicity (green – bottom right) include intrinsic sensitivity to radiation which may be related to SNPs or mutations in DNA damage/repair pathways including ATM, NBS1 and LIG4. A variety of genes validated as radiation-sensitive genes in other cancers have been identified as relevant to PRRT lung and gut sensitivity, that as they are. Furthermore, intrinsic radiation toxicity has been found for TNFα and TANC1 genes. None of these candidate genes have been evaluated for PRRT.