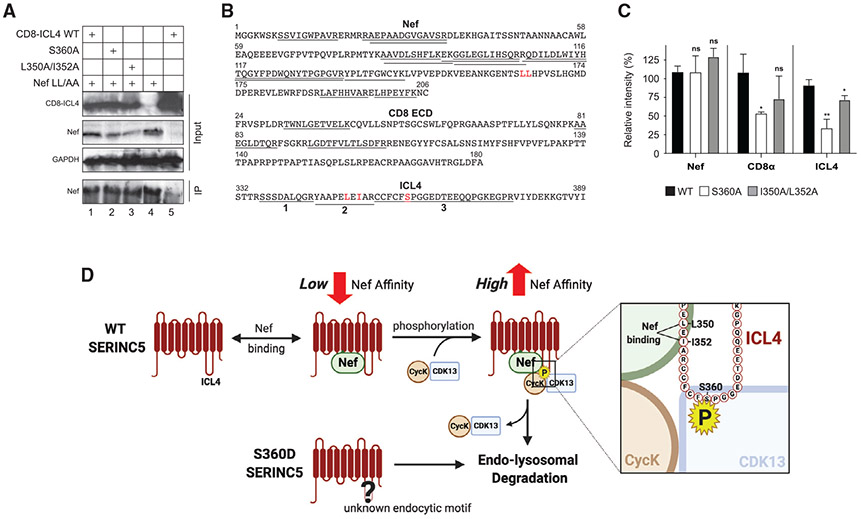
Figure 7. Detection of Nef and CD8-ICL4 interaction by quantitative proteomics.
(A) The indicated CD8-ICL4 fusion proteins were expressed with HIV-1 expressing Nef with a dileucine substitution (Nef LL/AA). Proteins were co-immunoprecipitated with a polyclonal anti-Nef antibody and detected by WB using the indicated antibodies.
(B) CoIP samples 1, 2, and 3 in (A) were analyzed by MS after trypsin digestion. Nef, CD8, and ICL4 peptides detected from these samples are underlined. Three ICL4 peptides are numbered (1, 2, 3).
(C) Levels of Nef, CD8, and ICL4 peptides detected by MS in (B) were quantified by label-free quantification (LFQ) and their intensity is presented as relative values, with the value of WT CD8-ICL4 set to 100%. Nef and CD8 were quantified by all detectable peptides and ICL4 was quantified by peptide-1 (SSSDALQGR). The error bars indicate SEM calculated from three independent experiments. Statistical analysis: *p < 0.05, **p < 0.01; ns, p > 0.05.
(D) A proposed model of how S360 phosphorylation promotes SERINC5 downregulation.