Figure 3.
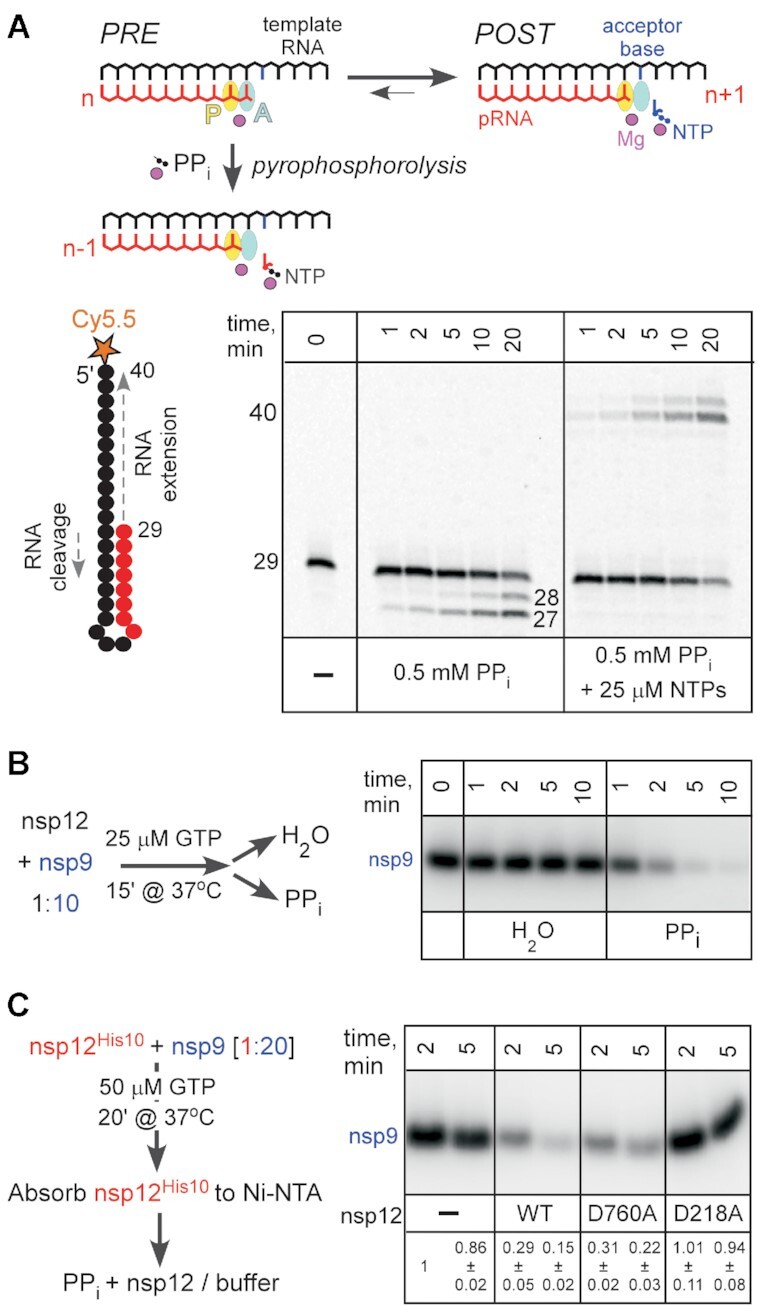
Removal of the GMP moiety in the presence of pyrophosphate. (A) Top: Pyrophosphorolysis of the nascent product RNA (pRNA; red). The active site (marked by the position of the catalytic Mg2+; magenta sphere) consists of two sub-sites: the P-site (product; yellow) and the A-site (acceptor; cyan). Following nucleotide addition, the pRNA 3′ end is bound in the A-site in the pre-translocated state. Upon forward translocation, the 3′ end moves to the P-site and the incoming substrate NTP (blue) can bind to the A-site through base pairing with the acceptor base of the RNA template strand (black). The pre-translocated state is sensitive to pyrophosphorolysis, which generates the NTP product and a one-nt shortened pRNA. Bottom: SARS-CoV-2 RdRp transcription complex assembled on the 5′ Cy5.5-labeled hairpin, which comprises both the template and the product RNAs, is completely resistant to PPi even in the absence of NTPs. (B) GMP removal from nsp9 in the presence of PPi. (C) The hand-over assay in which pre-GMPylated nsp9 is incubated with WT or mutant nsp12 variants. Signal intensity was compared to that observed with nsp9 incubated with buffer (set at 1) and is shown as mean ± SD (n = 3).
