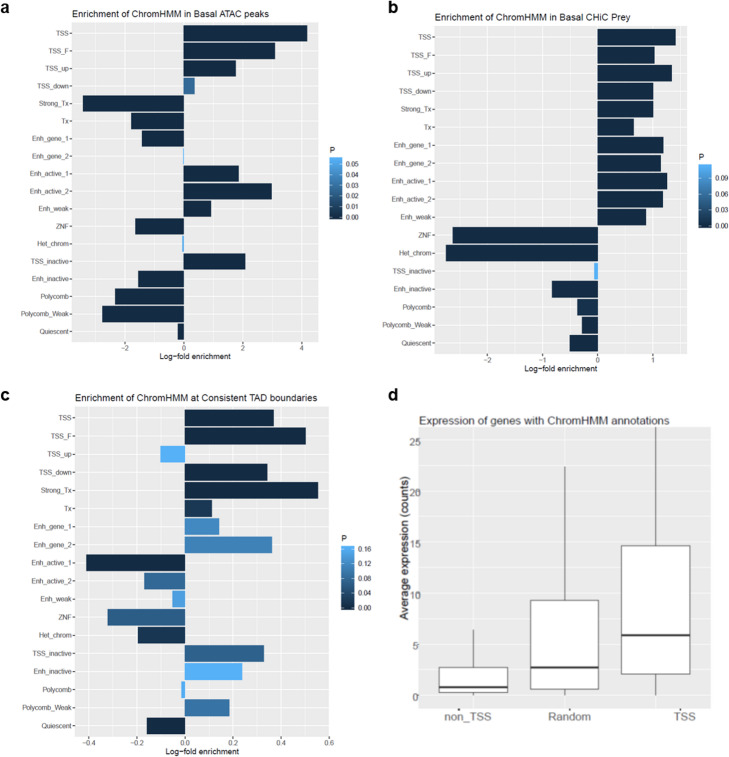
Fig. 2.
Cross validation of generated datasets defining the chromatin landscape of FLS. a Log fold change enrichment of chromatin states as defined by ChromHMM in open chromatin regions as identified by ATAC-seq. b Log fold change enrichment of chromatin states as defined by ChromHMM in prey fragments of Capture HiC measurements. c Log fold change enrichment of chromatin states as defined by ChomHMM in consistent TAD boundaries. d Basal average expression of genes (RNA-seq counts) across non-TSS, TSS, and random ChromHMM annotations. TSS = transcription start site, TSS_F = flanking TSS; TSS_up = upstream TSS; TSS_down = downstream TSS; Tx = Transcription; Enh_gene = enhancer genic; ZNF = zinc finger; Het_chrom = heterochromatin