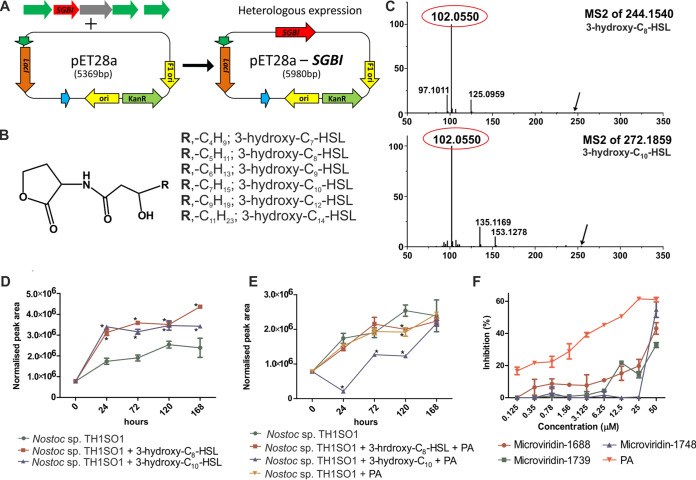
FIG 2.
(A) Heterologous expression of SGBI in BL21(DE3)/pET28a. (B) General structure of all the variants of HSL detected from the extract of heterologously expressed SGBI in BL21(DE3)/pET28a. (C) HRMS/MS product ion spectra of the two most abundant protonated molecules ([M + H]+) at m/z 244.1540 (3-hydroxy-C8-HSL) and m/z 272.1859 (3-hydroxy-C10-HSL) derived from the extract of heterologously expressed SGBI in BL21(DE3)/pET28a. The characteristic product ion at m/z 102.0550, corresponding to the deacylated homoserine lactone, was detected. (D) Induction of microviridin-1688 production after feeding with 3-hydroxy-C8-HSL/3-hydroxy-C10-HSL (at a 2.5 μM final concentration). The liquid chromatography-mass spectrometry (LC-MS) peak area was normalized to the dry biomass. (E) Inhibition of microviridin-1688 production in the presence of a quorum-sensing inhibitor, penicillic acid (PA), in combination with 3-hydroxy-C8-HSL/3-hydroxy-C10-HSL (at a 2.5 μM final concentration). (F) Dose-dependent inhibition activity of microviridin-1688, microviridin-1739, microviridin-1748, and penicillic acid on the QS-dependent bioluminescence of the lasR-based bioreporter strain E. coli/pSB1075 induced by its cognate molecule 3-oxo-C10-HSL at a noninhibitory concentration. The average bioluminescence observed for the negative control is used to calculate the relative inhibition percentage. Data are expressed as standard deviations (SD) of the means (n = 3). *, P < 0.001 versus the control by analysis of variance (ANOVA) followed by a Bonferroni posttest.