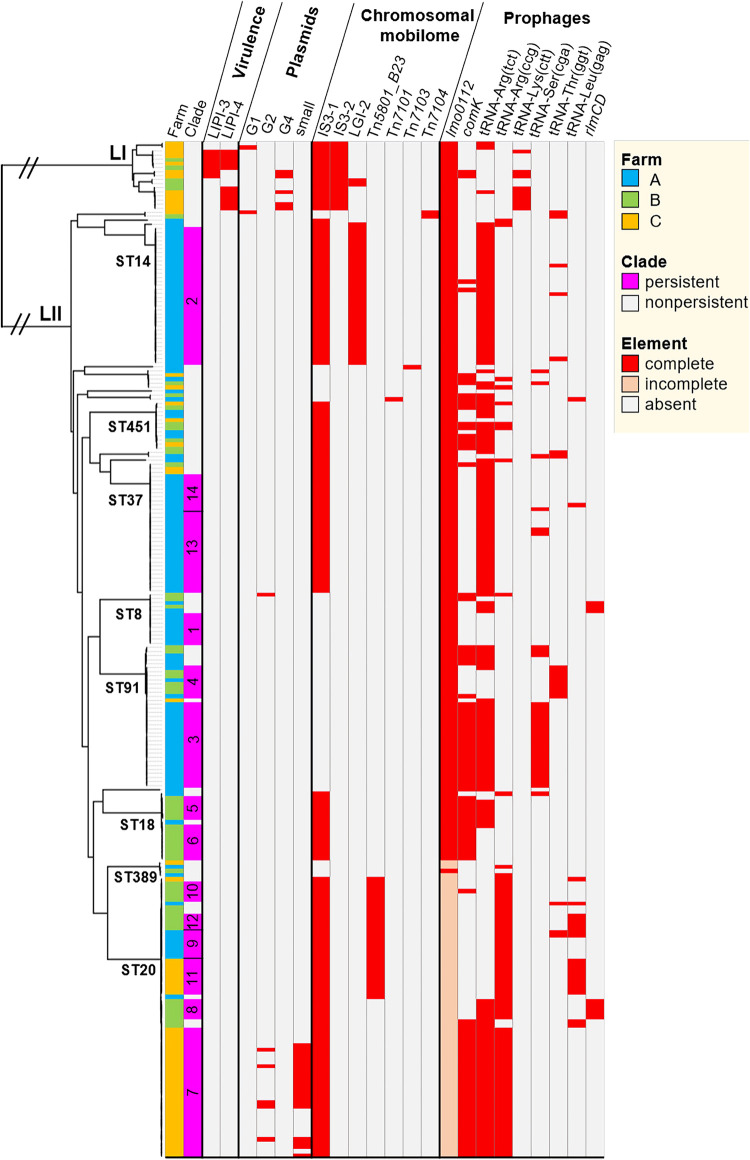
FIG 2.
Phylogeny and genomic elements of 250 L. monocytogenes dairy farm isolates. The Lyve-SET 1.1.4f single-nucleotide polymorphism (SNP)-calling pipeline was used to generate an alignment file of the 250 genomes using L. monocytogenes EGD-e (GenBank accession number NC_003210.1) as a reference. Recombinant sites were removed from the alignment using Gubbins 3.0. Maximum-likelihood phylogeny was inferred from concatenated SNP alignment files using PhyML 3.3. The tree was visualized using FigTree 1.4.4. Pathogenicity islands, plasmids, chromosomally located mobile elements, and prophages were identified from the assembled and annotated draft genomes. The heatmap is restricted to genomic elements that were detected in this study. Persistent clade numbers corresponding to data in Table 1 are shown. Plasmids are categorized by phylogenetic group and prophages by insertion site. L, lineage; ST, multilocus sequence type; LIPI, Listeria pathogenicity island; IS3, Listeria IS3-like element; LGI-2, Listeria genomic island 2.