FIG 4.
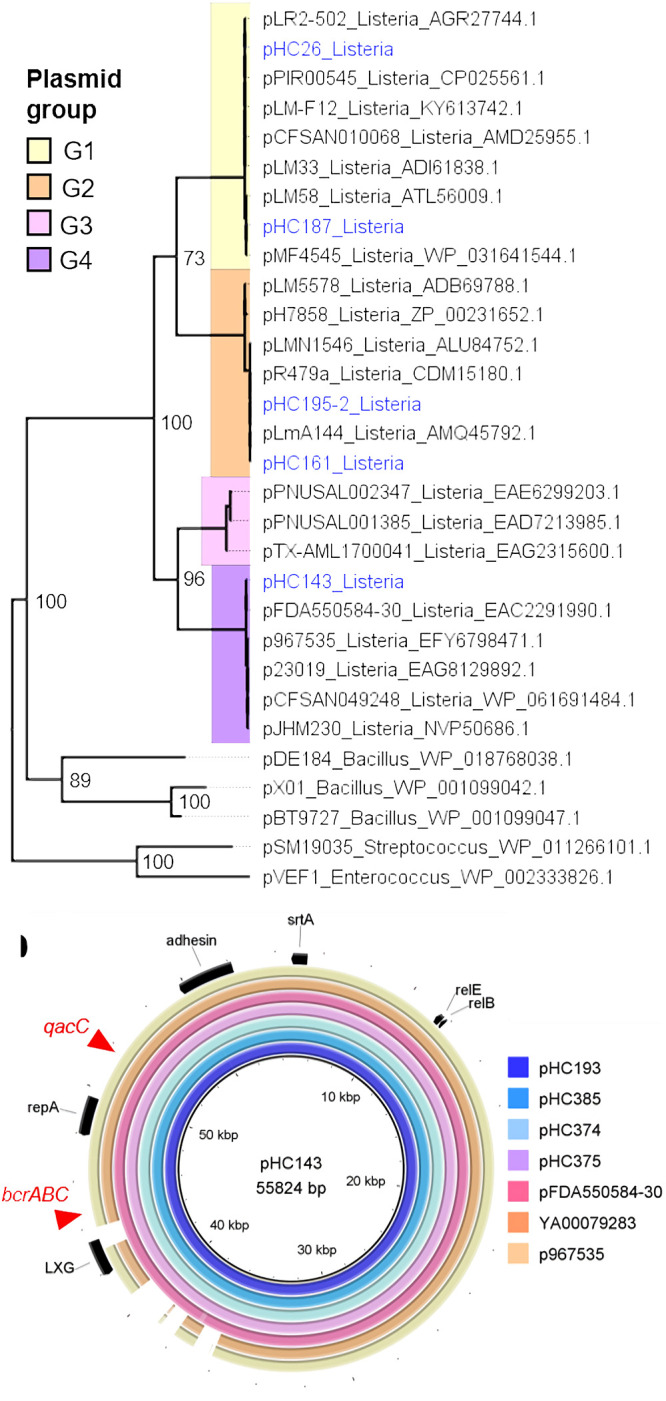
Characterization of plasmids based on RepA. (a) Maximum-likelihood phylogenetic analysis of the >50-kb plasmids detected in the present study, based on the repA amino acid sequences. The analysis employed the Jones-Taylor-Thornton substitution model with 100 bootstraps and was performed using MEGA7 software. Bootstrap support values above 70 are shown. Plasmids represented three phylogenetic clades (plasmid groups G1, G2, and G4). Plasmid groups correspond to the groups established by Kuenne et al. (28) and Schmitz-Esser et al. (29). Tip labels correspond to plasmid names and host genera; plasmids from this study are labeled in blue. (b) G4 plasmids of the L. monocytogenes strains HC193 (this study), HC374 (this study), and FDA550584-30 (BioSample accession number SAMN02923676) aligned with >95% identity across the entire length of pHC143 from this study; plasmids of the L. monocytogenes strains 967535 (BioSample accession number SAMN15680309) and YA00079283 (accession number SAMN08970420) aligned with >95% identity to most of pHC143. Red arrows indicate the insertion sites of the biocide resistance loci qacC, present in p967535 and pYA00079283, and bcrABC, present in pFDA550584-30. The alignment was generated using BRIG 0.95. For pHC143, plasmid length in base pairs (bp) is given.
