FIG 5.
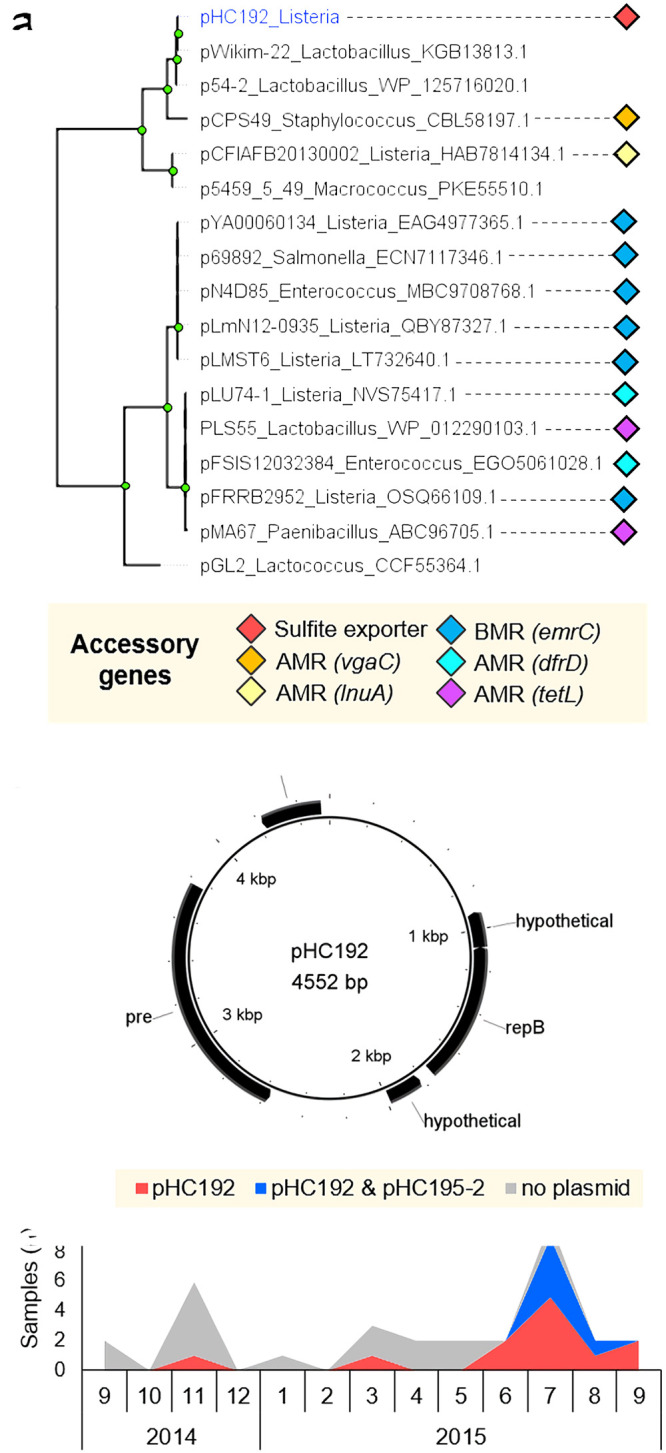
Phylogeny, gene content, and epidemiology of the novel plasmid pHC192. (a) Maximum-likelihood phylogenetic analysis of pHC192 and related plasmids, based on the repB amino acid sequences. Plasmids other than pHC192 were identified and obtained from GenBank using BLASTp. The analysis employed the Jones-Taylor-Thornton substitution model with 100 bootstraps and was performed using the MEGA7 software. Node labels indicate bootstrap support values above 70. Tip labels correspond to plasmid names and host genera; plasmids from this study are labeled in blue. Tip shapes depict harborage of resistance genes against antimicrobials (antimicrobial resistance [AMR]) and biocides (biocide resistance [BCR]). (b) The 4.5-kb plasmid pHC192, carrying a putative SafE/TauE family sulfite exporter (GenBank accession number WP_016896343.1). The figure was constructed using BRIG 0.95. Plasmid length in base pairs (bp) is given. (c) Numbers of samples containing no plasmid, pHC192, or both pHC192 and pHC195-2 among persistent clade C7 isolates during each month of sampling. Plasmid prevalence in C7 isolates increased over the 1-year sampling period.
