Figure 7.
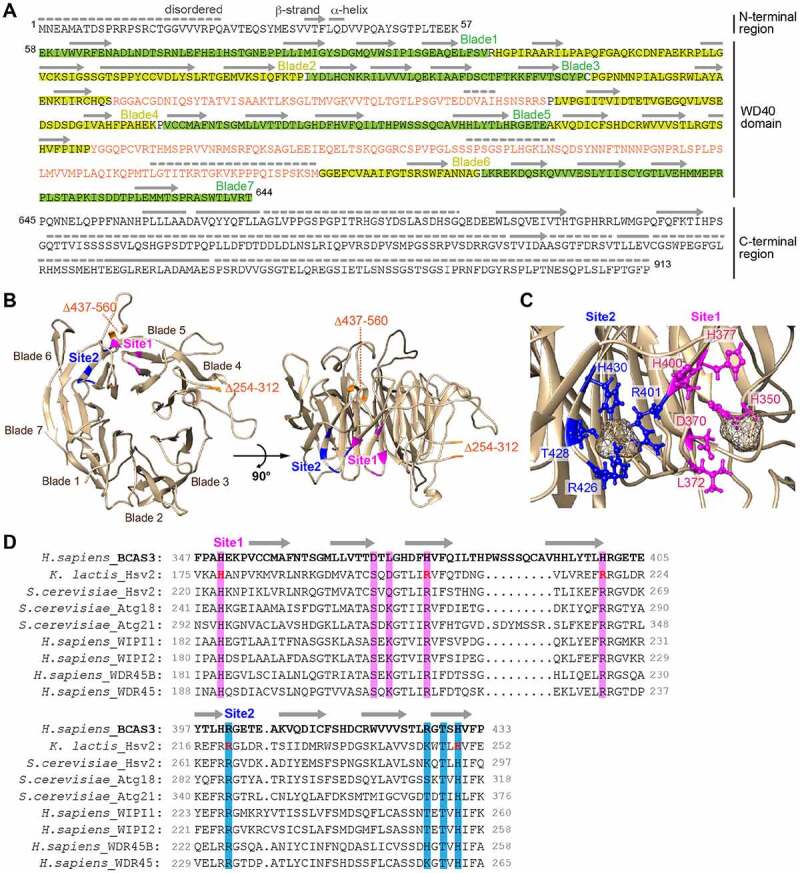
Two phospholipid-binding sites are predicted in the BCAS3 WD40 repeat domain. (A) Predicted secondary structure for human BCAS3. The secondary structure in the WD40 repeat domain was assigned based on the model shown in (B) and secondary structure features in the N- and C-terminal regions were determined using PSIPRED [76]. Disordered regions were predicted using DISOPRED3 [77]. Lines, arrows, and dotted lines indicate helix, strand and disordered region, respectively. The two inserts in blades 4 and 6 are colored orange. (B) Predicted 3D structure of the human BCAS3 WD40 repeat domain. The two inserts in WD40 (i.e. residues 254–312 and 437–560) are deleted. Sites 1 and 2 (the predicted phospholipid-binding pockets) are colored magenta and blue, respectively. (C) Detailed sulfate binding depicted for sites 1 and 2. Side chains of the indicated residues in site 1 (magenta) and site 2 (blue) are depicted in ball-and-stick mode, whereas the sulfate ions are depicted as van der Waals spheres. (D) Alignment of the amino acid sequence around sites 1 and 2 of human BCAS3 and various Atg18 family proteins. Residues critical for phospholipid binding in K. lactis Hsv2 [63] are colored in red. Amino acid residues in sites 1 and 2 that were functionally examined in Figure 8 are highlighted in magenta and blue, respectively
