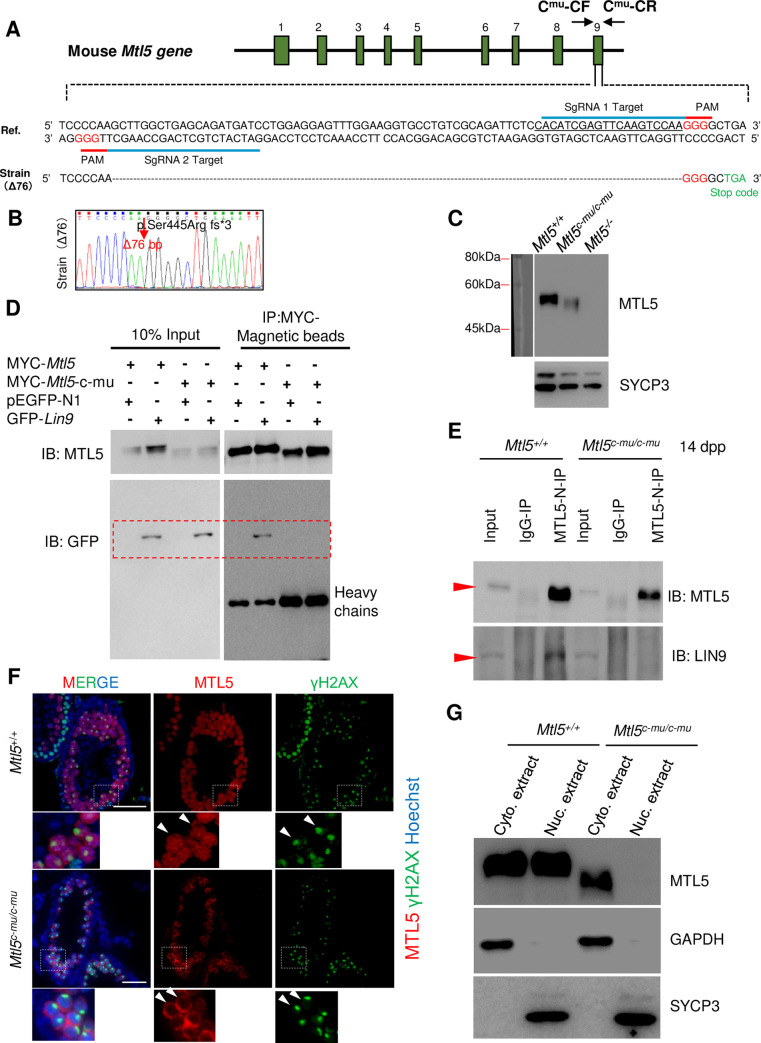
Fig 5. The C-terminus deleted MTL5 (Ser445Arg fs*3) exhibits abolished interaction with LIN9 and fails to be translocated into nuclei of spermatocytes in Mtl5c-mu/c-mu mice.
(A) Diagram (top) and nucleotide sequence (bottom) of the mouse Mtl5 locus from the Ensemble database (ENSMUST00000025840.15) with highlighted coding exons (green) and the TAG (light green) premature stop codon of the modified transcript (bottom). PAM, protospacer adjacent motif. sgRNA, single-guide RNA. Cmu-CF and Cmu-CR represent the genotyping primers of Mtl5 mutant mice. (B) The mutant mouse strain with a 76 bp deletion from position 1335 to 1410 in exon 9 (Mtl5c-mu/c-mu) was confirmed by Sanger sequencing. The position of the deletion site in the truncated MTL5 (p.Ser445Arg fs*3) produced by Mtl5c-mu/c-mu is indicated by the arrow. (C) Western blot analysis of MTL5 levels in testes from Mtl5+/+, Mtl5c-mu/c-mu, and Mtl5-/- mice. SYCP3 served as loading control to indicate the comparable composition of cell populations. (D) Western blot analysis of the interaction between truncated MTL5 (p.Ser445Arg fs*3) and LIN9 in HEK293T cells after co-transfection followed by co-immunoprecipitation with MYC-Magnetic beads. MYC-Mtl5-c-mu encoding the truncated MTL5 (p.Ser445Arg fs*3). pEGFP-N1 served as the negative control. IP, immunoprecipitation. IB, immunoblotting. (E) Western blot analysis of the interaction between truncated MTL5 (p.Ser445Arg fs*3) and LIN9 in 14 dpp Mtl5+/+ and Mtl5c-mu/c-mu mouse testes after co-immunoprecipitation with MTL5-N antibody. IP, immunoprecipitation. IB, immunoblotting. Red arrowheads indicate the expected bands. (F) Immunostaining of MTL5 (red) and γH2AX (green) on testicular sections from 4-week-old Mtl5+/+ and Mtl5c-mu/c-mu mice. Nuclei were counterstained with Hoechst 33342 (blue). White arrowheads indicate the pachytene spermatocytes with intact sex-body. Scale bars, 50 μm. (G) Western blot analyses of MTL5 and truncated MTL5 (p.Ser445Arg fs*3) using cytoplasmic and nuclear protein extracts from 4-week-old Mtl5+/+ and Mtl5c-mu/c-mu mouse testes. GAPDH and SYCP3 served as cytoplasmic and nuclear controls, respectively.