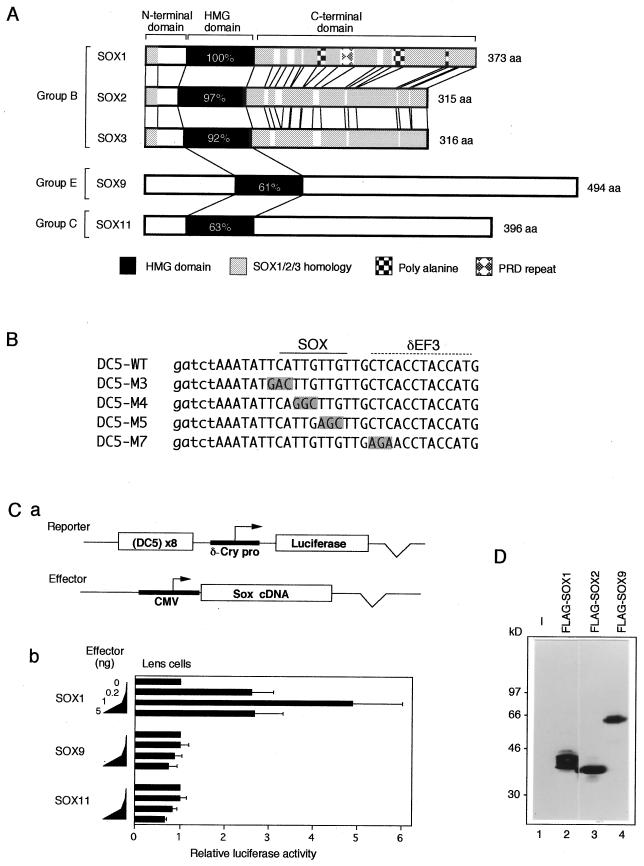
FIG. 1.
Comparison of SOX1/2/3, SOX9, and SOX11 with respect to protein structure and activation of the DC5 enhancer. (A) Schematic presentation of chicken SOX proteins used in this study. Percentages of amino acid identity with SOX1 in the HMG domain are given. (B) Sequences of the DC5 fragments used in this study. In the DC5 mutant sequences (22), altered bases are shaded. The SOX binding site and the putative δEF3 site are indicated at the top. Lowercase letters indicate bases introduced to generate BglII and BamHI restriction sites. (C) (a) Scheme of the cotransfection assay. The luciferase reporter plasmid contains octamerized DC5 fragment positioned upstream of δ1-crystallin minimal promoter (δ-Cry pro; −51 to +57). The cDNAs of Sox genes were expressed by the CMV enhancer-promoter. (b) Comparison of effects of SOX1, SOX9, and SOX11 on the DC5 enhancer. Lens cells were transfected with the luciferase reporter plasmid and with various amounts of effector vectors (0, 0.2, 1, and 5 ng) encoding one of the SOX proteins. Luciferase activity generated by the reporter in the absence of exogenous SOX was taken as 1. (D) Expression of FLAG-tagged SOX proteins in lens cells. Lens cells were transfected with pCMV/SV-Flag1 plasmids encoding no protein (−) or the indicated SOX proteins. Whole-cell lysates were subjected to Western blot analysis using anti-FLAG antibody. Calculated molecular masses of FLAG-SOX proteins are as follows: FLAG-SOX1, 39.2 kDa; FLAG-SOX2, 35.8 kDa; and FLAG-SOX9, 56.4 kDa. The sizes of molecular mass markers are indicated on the left.