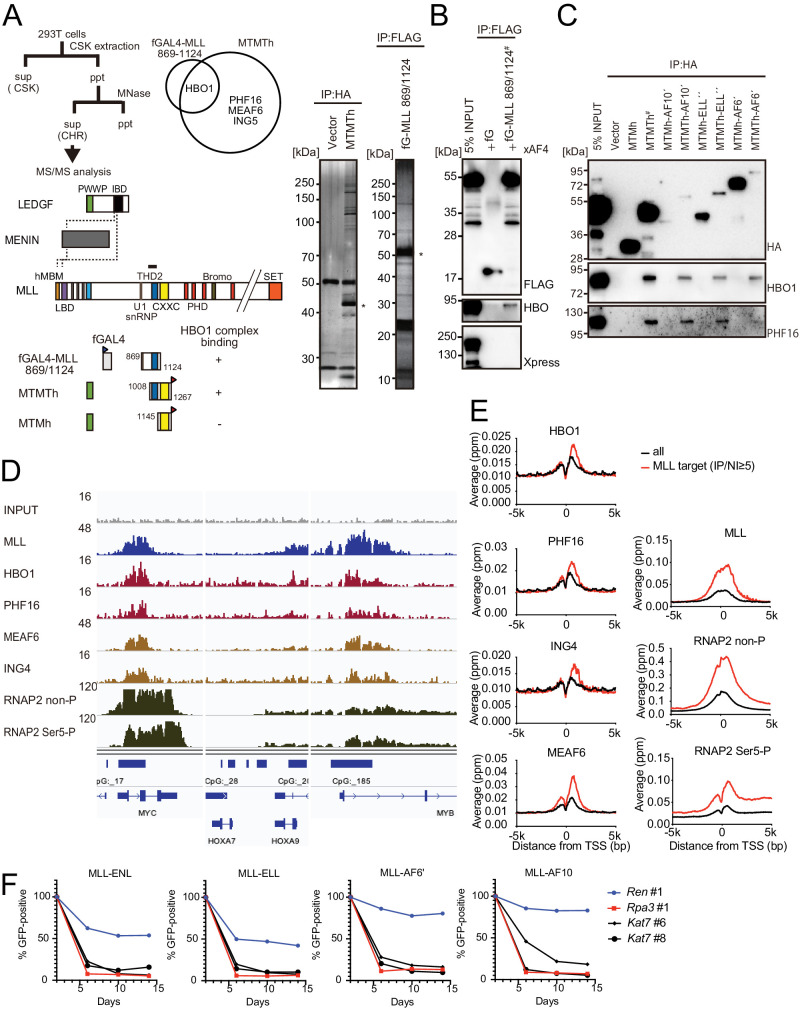
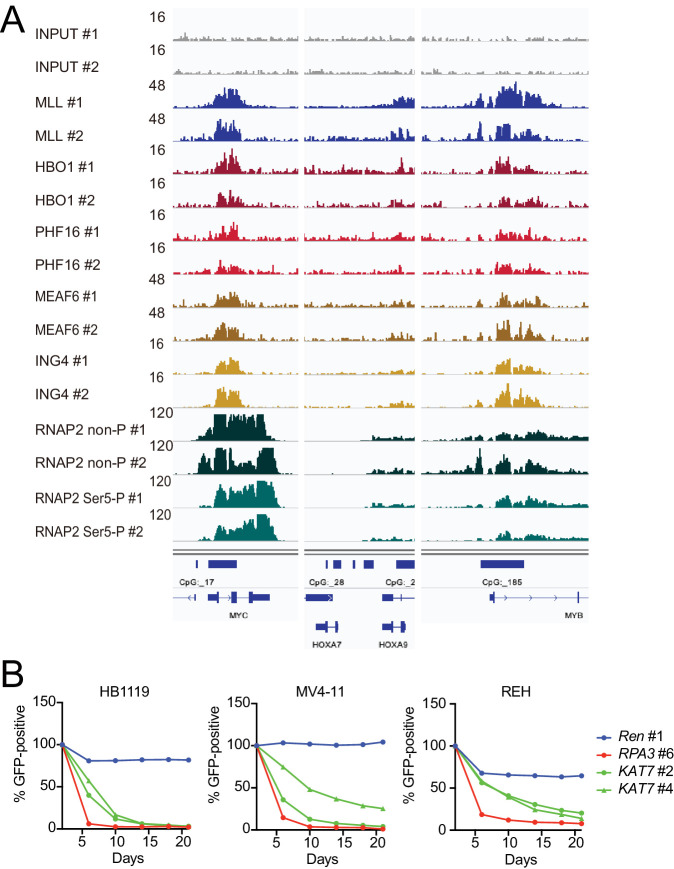
Figure 2. HBO1 complex associates with MLL proteins at promoters.
(A) Purification of trithorax homology domain 2 (THD2) domain-associating factors. A FLAG-tagged (f: indicated as a blue triangle) GAL4 DNA-binding domain fused with the MLL fragment containing the residues 869–1124 or HA-tagged MTMT fragment was transiently expressed in HEK293T cells. A schema of the fractionation-assisted chromatin immunoprecipitation (fanChIP) method is shown on top. The transgene products were purified from the chromatin fraction and analyzed by mass spectrometry. Silver-stained images (right) of the purified materials are shown. Asterisk indicates the position of the transgene products. A Venn diagram of identified THD2 domain-associating factors by mass spectrometry is shown. (B) Association of GAL4-THD2 fusion with HBO1. Immunoprecipitation (IP)-western blotting of the chromatin fraction of HEK293T cells transiently expressing FLAG-tagged GAL4-MLL 869/1124 construct and Xpress-tagged AF4 (xAF4) was performed. Co-purification of HBO1, but not xAF4, was confirmed. #: the sample used for the input. (C) THD2-dependent association with the HBO1 complex. IP-western blotting of the chromatin fraction of virus-packaging cells, transiently expressing various HA-tagged MTMT (or MTM) fusion constructs, was performed. (D) Genomic localization of MLL and the HBO1 complex components in HB1119 cells. The ChIP followed by deep sequencing (ChIP-seq) profiles were visualized using the Integrative Genomics Viewer (The Broad Institute). The minimum value of the y-axis was set at 0, while the maximum value for each sample is indicated. (E) Average distribution of proteins near the transcription start sites (TSSs) of HB1119 cells. Genes whose MLL ChIP signal/input ratio at the promoter proximal transcribed region was ≥5 were defined as MLL target genes. Average ChIP signal distribution of indicated proteins at the MLL target genes (red) or all genes (black) is shown. The y-axis indicates the frequency of the ChIP-seq tag count (ppm) in 25 bp increments. (F) Requirement of HBO1 for myeloid progenitors immortalized by various oncogenes. sgRNA competition assays for Hbo1 were performed on immortalized myeloid progenitors. The ratio of GFP-positive cells co-expressing sgRNA was measured by flow cytometry. sgRNA for Renilla luciferase (Ren) was used as a negative control, which does not affect proliferation. sgRNA for Rpa3 was used as a positive control, which impairs proliferation.