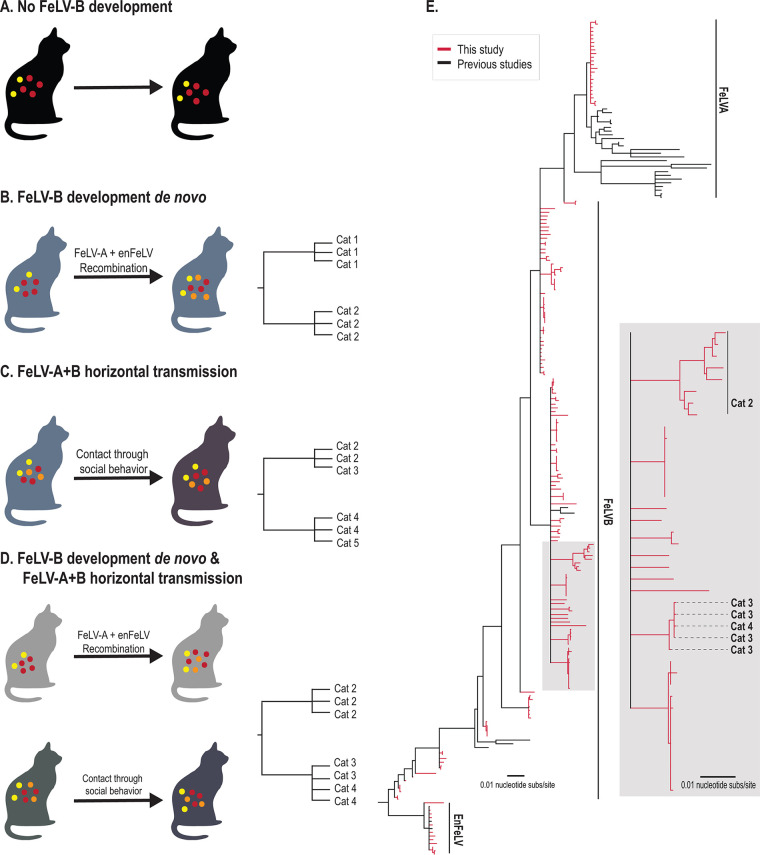
FIG 1.
FeLV-B phylogeny reveals a high degree of genetic dissimilarity within the closed cat colony and supports de novo recombination as a primary source of FeLV-B emergence. (A to D) Four potential scenarios for FeLV-B emergence were considered, which could be distinguished by phylogenetic analysis. All members of the Felis genus harbor enFeLV (yellow circles as shown for each scenario). (A) No recombination. Active FeLV-A infection (red circles) does not develop into FeLV-B, despite the cat harboring enFeLV (yellow circles). Ten of 32 cats evaluated were in this category. (B) A cat has an active FeLV-A infection (red circles) that recombines with enFeLV (yellow circles) to develop an FeLV-B (orange) infection. If all FeLV-B infections developed de novo and horizontal transmission was not possible or rare, genomic analysis would cluster viral sequences from the same individual together or show recombination events that are not identical between individuals. (C) A cat horizontally transmits both FeLV-A (red circles) and FeLV-B (orange circles) to another cat via social contact. In this case, phylogenies indicate branches with shared FeLV-B ancestry between infected individuals. (D) FeLV-B infection (orange circles) emerges via both de novo recombination and horizontal transmission. Resulting phylogeny would demonstrate both independent FeLV-B branches related to each cat as well as mixed branches indicating FeLV-B infections shared between individuals. It is important to note that different FeLV-B recombination break points that occur within an individual could be shared across individuals and would generate a phylogenic structure similar to that of direct transmission of FeLV-B and could confound interpretations. (E) Neighbor-joining phylogenetic tree illustrates the relationship between FeLV sequences (FeLV-A and -B and enFeLV) recovered in this study (red) and sequences reported previously (black). Two primary clades and several minor clades were identified. Several FeLV-B clades are seen here, which are indicative of the differing recombination patterns between FeLV-A and enFeLV. This results in FeLV-B isolates that are more closely related to FeLV-A, isolates that are more enFeLV-like, or variants that are intermediate between these two identities. Our data suggest scenario D, although the majority of FeLV-B variants arise de novo.