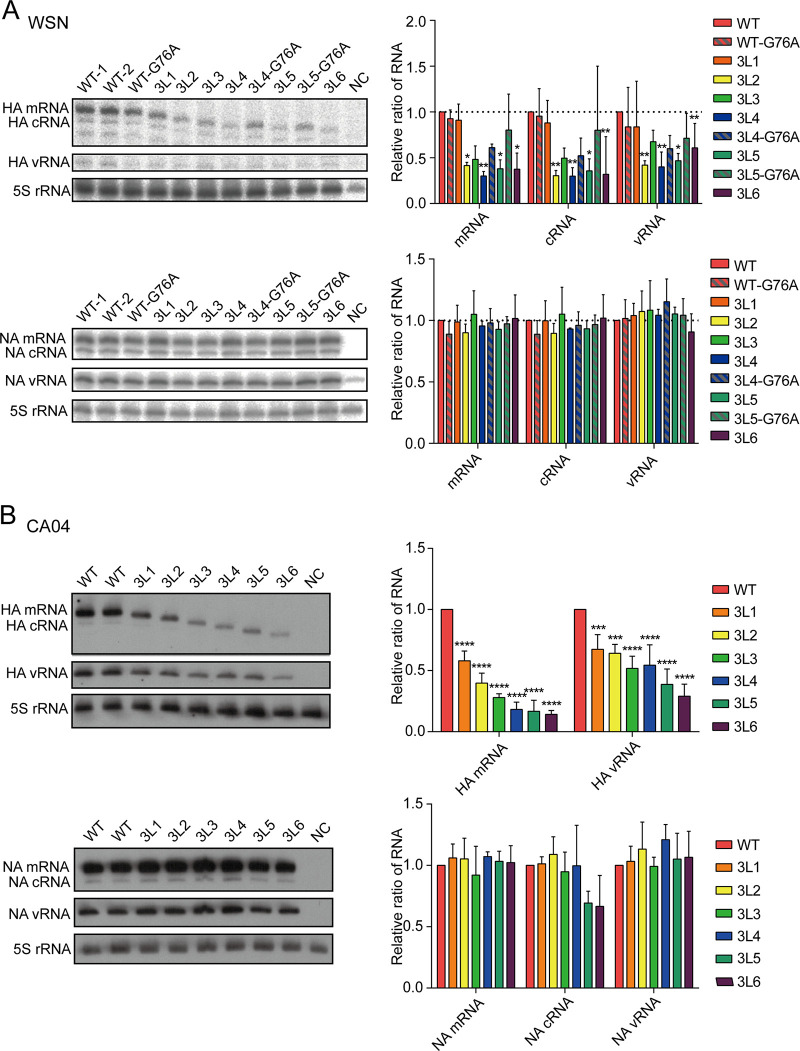
FIG 5.
The 3′-end H1-ssNCR-truncated mutant templates are replicated less efficiently than the wild-type HA template in multiple-template RNP reconstitution assays. (A, B) Seven pHW2000-PB2, -PB1, -PA, -HA (wild type or mutant), -NP, -NA, and -NS plasmids derived from WSN (A) or CA04 (B) were transfected into HEK-293T cells. A polymerase containing an active site mutation (PB1a, D445A/446A) was used as negative control (NC). Accumulation of HA and NA RNAs was analyzed by primer extension and 6% PAGE. The graph shows the mean intensity signals of mutant HA and NA RNAs relative to those of wild-type RNAs. The data represent the means ± SEM of three independent experiments. Two-way analysis of variance (ANOVA) with Dunnett correction for multiple testing; asterisks represent a significant difference from wild-type virus as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.