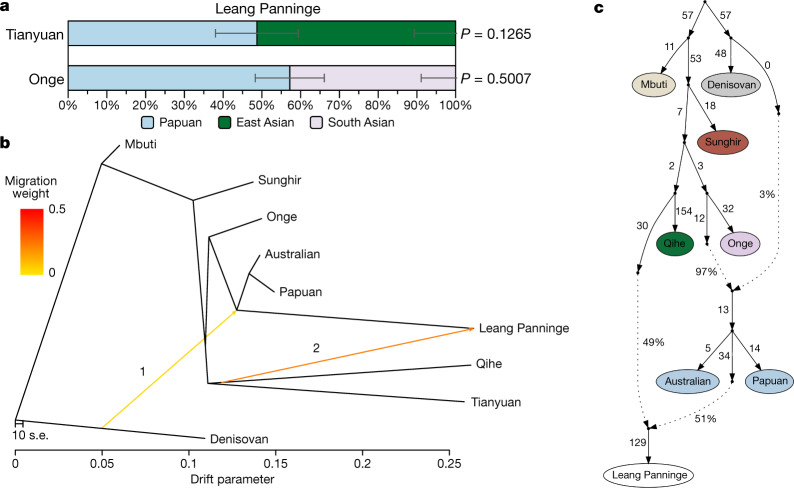
Fig. 3. Admixture signals detected in the Leang Panninge genome.
a, Admixture proportions modelling Leang Panninge as a combination of Papuan49 and Tianyuan38 or Onge49 groups as estimated by qpAdm33 using Mbuti, Denisovan39, Kostenki 14 (ref. 50) and ancient Asian individuals1,37 as rotating reference groups (Supplementary Table 26). The error bars denote standard errors as calculated with block jacknife in the qpAdm software. b, c, Admixture graphs placing Leang Panninge on the branch with the present-day Near Oceanian clade41 and showing the admixture with a deep Asian-related ancestry in TreeMix42 (b) (Extended Data Fig. 10, Supplementary Fig. 6) and qpGraph (c) (worst z-score of −2.194; Supplementary Figs. 7–11)33,37–39,43. In b, ‘1’ and ‘2’ refer to the order in which the TreeMix software added ‘migration events’ (indicated by the arrows) to the graph. When plotting qpGraph results (c), the dotted arrows indicate admixture edges.