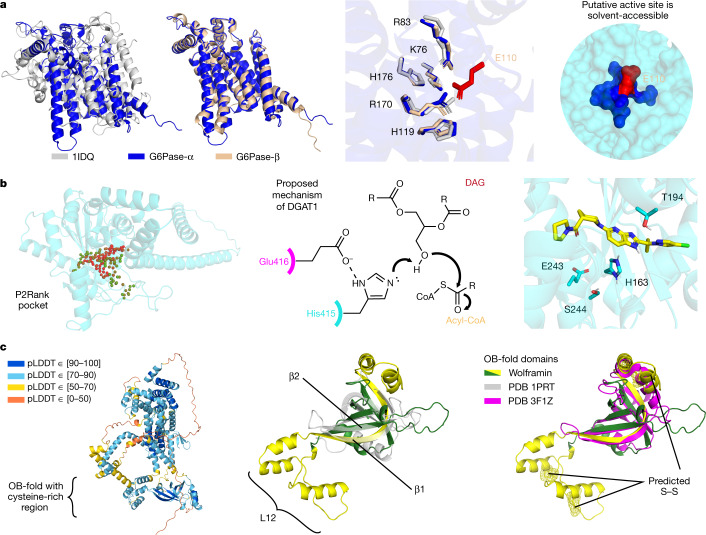
Fig. 3. Highlighted structure predictions.
a, Left, comparison of the active sites of two G6Pases (G6Pase-α and G6Pase-β) and a chloroperoxidase (PDB 1IDQ). The G6Pases are glucose-forming enzymes that contain a conserved, solvent-accessible glutamate (red; right) opposite the shared active-site residues (middle). b, Left, pocket prediction (P2Rank65) identifies a putative binding pocket for DGAT2, which is involved in body-fat synthesis. Red and green spheres represent the ligandability scores by P2Rank of 1 and 0, respectively. Middle, a proposed mechanism for DGAT151 activates the substrate with Glu416 and His415, which have analogous residues in the DGAT2 pocket. The docked inhibitor is well placed for polar interactions with His163 and Thr194 (right). The chemical structure (middle) is adapted from ref. 51. c, Predicted structure of wolframin, mutations in which cause Wolfram syndrome. Although there are regions in wolframin with low pLDDT (left), we could identify an OB-fold region (green/yellow), with a comparable core to a prototypical OB-fold (grey; middle). However, the most similar PDB chain (magenta; right) lacks the conserved cysteine-rich region (yellow) of our prediction. This region forms the characteristic β1 strand and an extended L12 loop, and is predicted to contain three disulfide bridges (yellow mesh).