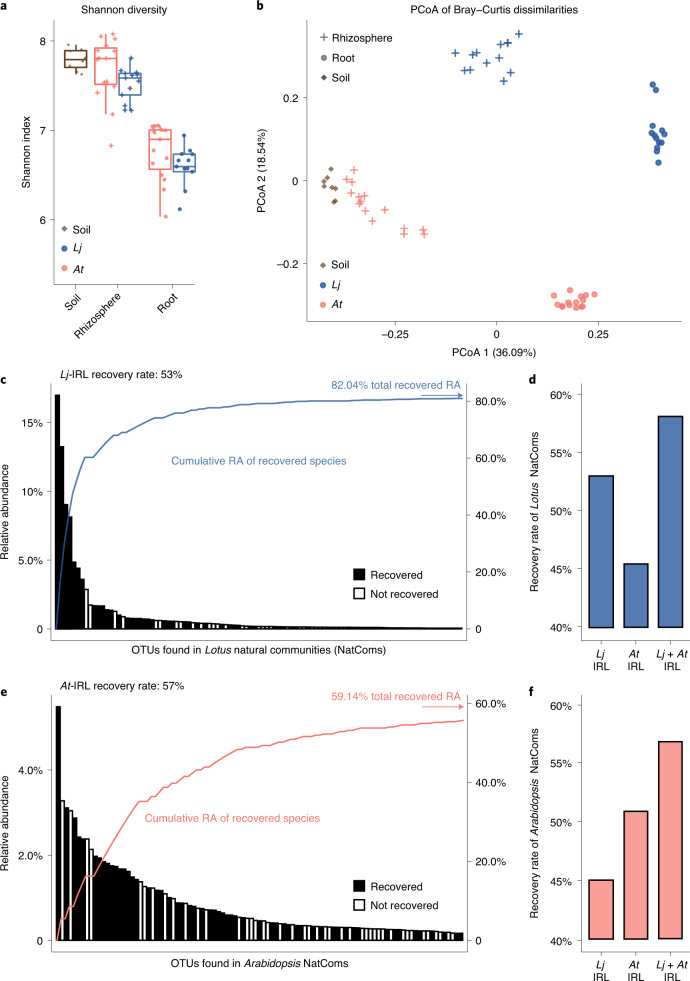
Fig. 1. Lotus and Arabidopsis root-associated bacterial communities.
a, Alpha diversity analysis of soil- (n = 8), rhizosphere- (n = 13 for Gifu, n = 15 for Col-0) and root-associated bacterial communities (n = 13 for Gifu, n = 15 for Col-0) from Lj and At plants grown in natural soil (exp. A), assessed using the Shannon index. b, PCoA of Bray–Curtis dissimilarities of the same communities (n = 64). c,e, Rank abundance plots of OTUs found in the Lotus (c) and Arabidopsis (e) natural root communities. Community members captured in the corresponding culture collection are depicted as black while non-recovered OTUs are shown in white. The vertical axis on the right shows the accumulated relative abundance in natural communities of all recovered OTUs. d,f, Percentage of abundant OTUs (≥0.1% RA) associated with Lotus (d) or Arabidopsis (f) roots in nature (natural communities, NatComs) that are captured in the Lotus or the Arabidopsis IRLs (At- and Lj-IRL).