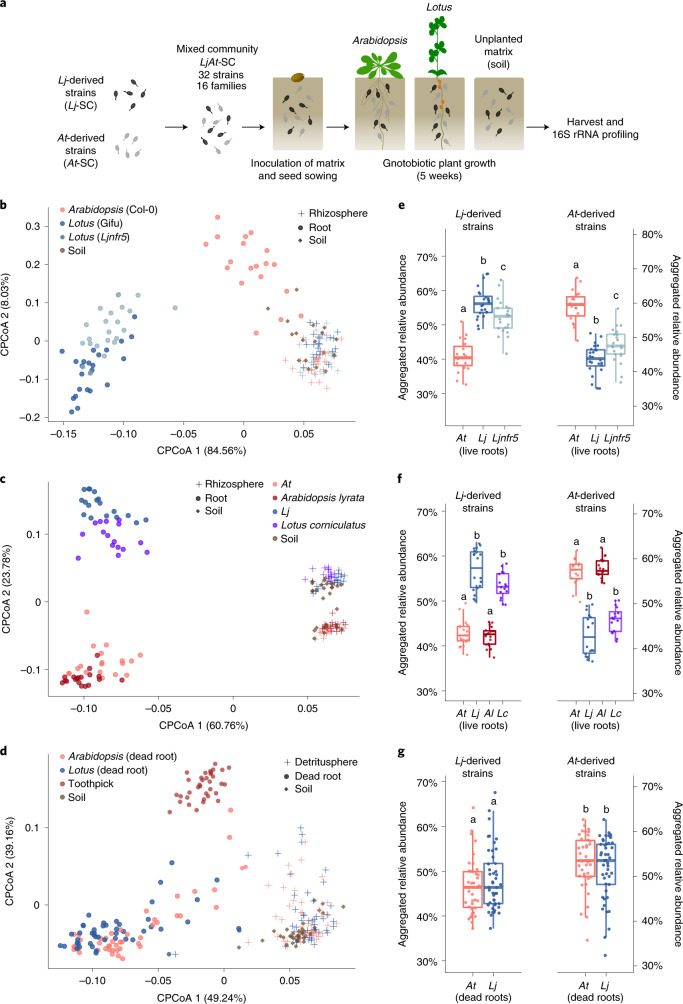
Fig. 3. Reconstitution experiments recapitulate culture-independent patterns and show signatures of host preference by commensal communities.
a, Setup of the competition experiments. b–d, Constrained PCoA (CPCoA) of Bray–Curtis dissimilarities (constrained by all biological factors and conditioned by all technical variables) of soil, rhizosphere and root samples. b, Lj wild-type Gifu, nfr5 mutant and At wild-type Col-0 plants cocultivated with the mixed community LjAt-SC2 (exp. B, n = 155, variance explained 53.8%, P = 0.001). c, Gifu, Col-0, A. lyrata MN47 (Al) and L. corniculatus cocultivated with LjAt-SC3 (exp. F, n = 173, variance explained 65.1%, P = 0.001). d, Dead roots of Gifu and Col-0, and toothpick cocultivated with LjAt-SC3 (exp. J, n = 250, variance explained 43.9%, P = 0.001). e–g, Aggregated RA of the 16 Lj-derived and the 16 At-derived strains in the live (e,f) or dead roots (g) of Lotus and Arabidopsis plants inoculated with LjAt-SC2 (n = 66, e) or LjAt-SC3 (n = 72, f and n = 89, g).