Figure 3.
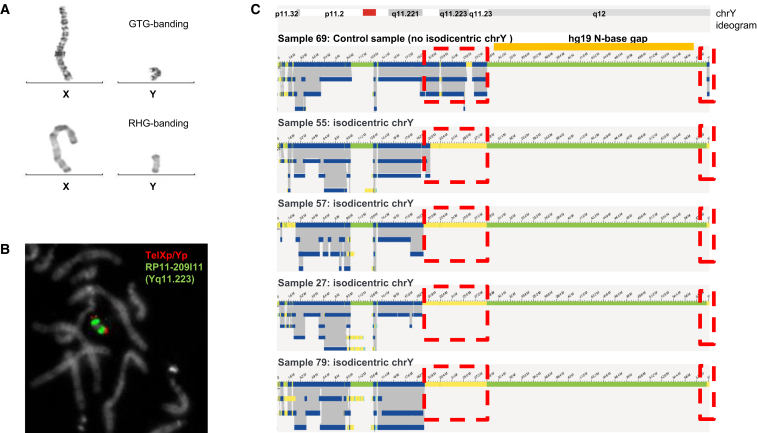
Isodicentric Y chromosomes show specific assembly map patterns
(A) G-banding with trypsin-Giemsa (GTG) and reverse banding using heat and Giemsa (RHG) of X and Y chromosomes of sample 57.
(B) FISH for sample 57 via fluorescently labeled probes TelXp/Yp (green) and RP11-209I11 (red, located at Yq11.223).
(C) Genome maps of Y chromosomes of samples 69 (normal chromosome Y), 55, 57, 27, and 79. Dotted red boxes indicate where isodicentric Y chromosomes have no coverage when compared to non-isodicentric Y chromosomes (the top genome map) corresponding to the deleted segment. Most likely the genome maps of the four isochromosomes by optical genome mapping point to three different breakpoints (deletion starts), (sample 55, 21.0 Mb; samples 57 and 79, 20.1 Mb; and sample 27, 19.9 Mb). The first three breakpoints coincide with palindrome 5 (hg19, chrY: 19,567,684–20,063,140 bp), which may be involved in the mediation of the isochromosomes or cause issues in a more precise assembly with the current optical genome mapping. The deletion in sample 55 locates in direct proximity of palindrome 4 (hg19, chrY: 20,612,099–20,802,247 bp).