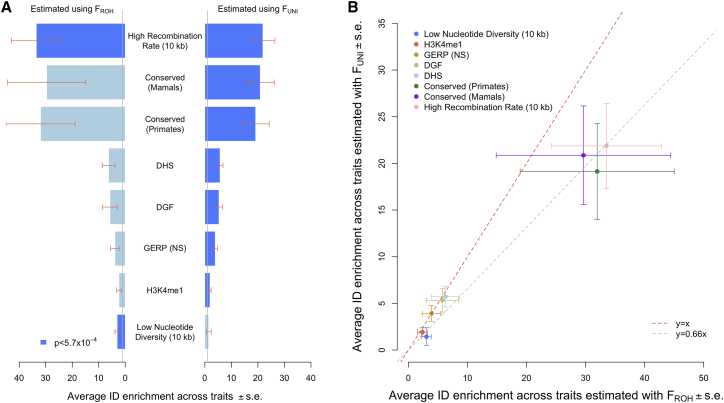
Figure 2.
Significant inbreeding depression (ID) enrichment within eight genomic annotations
(A and B) Estimates of ID enrichment on average across 11 traits obtained via and . Statistical significance was set at p < 0.05/(44 × 2) ≈ 5.7 × 10−4. Recombination rate and nucleotide diversity were analyzed as continuous annotations. “High recombination rate” denotes that recombination rate is positively correlated with ID, and “low nucleotide diversity” denotes that nucleotide diversity is negatively correlated with ID. Data underlying this figure are reported in Table S1. DHS, DNA-se I hypersensitive sites; DGF, chromatin accessible regions from digital genomic footprint; GERP (NS), GERP++ score number of substitution; H3K4Me1, histone mark (binary annotation). Error bars represent standard errors (SEs).