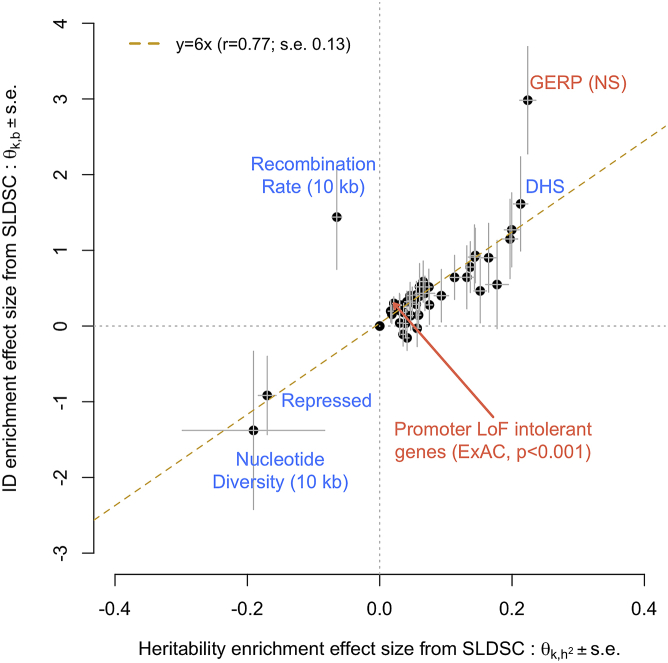
Figure 3.
Correlation between functional enrichment of SNP-based heritability and inbreeding depression (ID) estimated from GWAS summary statistics
Enrichment statistics for heritability (x axis: ) and ID (y axis: ) were estimated with stratified LD score regression (SLDSC) as described in the material and methods section. Enrichments statistics were averaged over 11 traits. Each dot represents a genomic annotation, and errors bars represent standard errors (SEs) calculated with a block-jackknife procedure. Annotations highlighted in coral font show a statistically significant ID enrichment at p < 0.05/88. DHS, DNA-se I hypersensitive sites; GERP (NS), GERP++ score number of substitution; LoF, loss of function. Data underlying this figure are reported in Table S5.