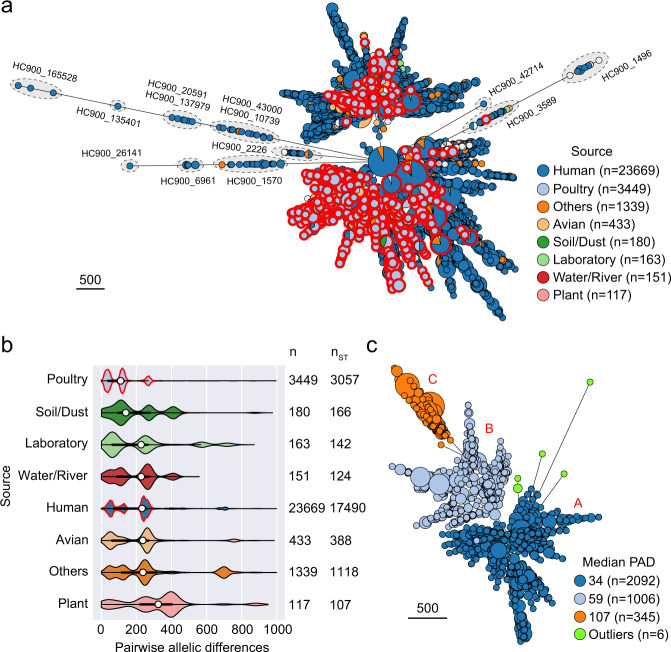
Fig. 3. Global population structure and genomic diversity of Salmonella Enteritidis.
a Minimum spanning tree of 30,015 Salmonella Enteritidis isolates of 22,775 cgMLST sequence types (cgSTs) in EnteroBase. Each node represents a distinct cgST or cgSTs differed by only missing data. The size of each node indicates the number of isolates within that node. Isolates are color coded by EnteroBase-defined sources. Only sources with at least 100 cgSTs are shown. Nodes containing poultry isolates are highlighted by red border. The majority of isolates belong to the HC900_12 cluster defined by EnteroBase. Clusters not belonging to HC900_12 are highlighted by dashed ovals. The scale bar indicates 500 cgMLST alleles. An interactive version of the tree is available at https://enterobase.warwick.ac.uk/ms_tree/51520. b Comparison of intra-source pairwise allelic differences (PADs) based on individual STs. Violin plots show probability densities of PADs. Each boxplot within a violin plot summarizes the distribution of PADs of all isolates in a source. White circles indicate the median PAD values. Thick black bars represent the interquartile ranges. Thin black lines indicate the ranges between maximum and minimum values. Only sources with numbers of isolates (n) and cgSTs (nST) ≥ 100 are shown. “Others” includes isolates without source information or not belonging to any specific source defined in EnteroBase. PADs larger than 1,000 are considered outliers and not included. The peaks of kernel density estimate of the PAD distributions of poultry and human isolates are outlined in red. c Minimum spanning tree of 3,449 Salmonella Enteritidis poultry isolates of 3,057 cgSTs in EnteroBase. Three sub-populations of poultry isolates (A, B, and C) and their individual median PADs are shown. Six outlier isolates (green) belong to four different HC200 clusters including HC200_13575 (n = 3), HC200_12160 (n = 1), HC200_1522 (n = 1). The rest of isolates in A, B, and C belong to the same HC200 cluster, HC200_12. The scale bar indicates 500 cgMLST alleles. An interactive version is available at https://enterobase.warwick.ac.uk/ms_tree/50727.