Figure 5. Ribosomal siRNAs (risiRNAs) accumulate in the germline of prg-1(−) mutant and depend on the endogenous RNAi machinery.
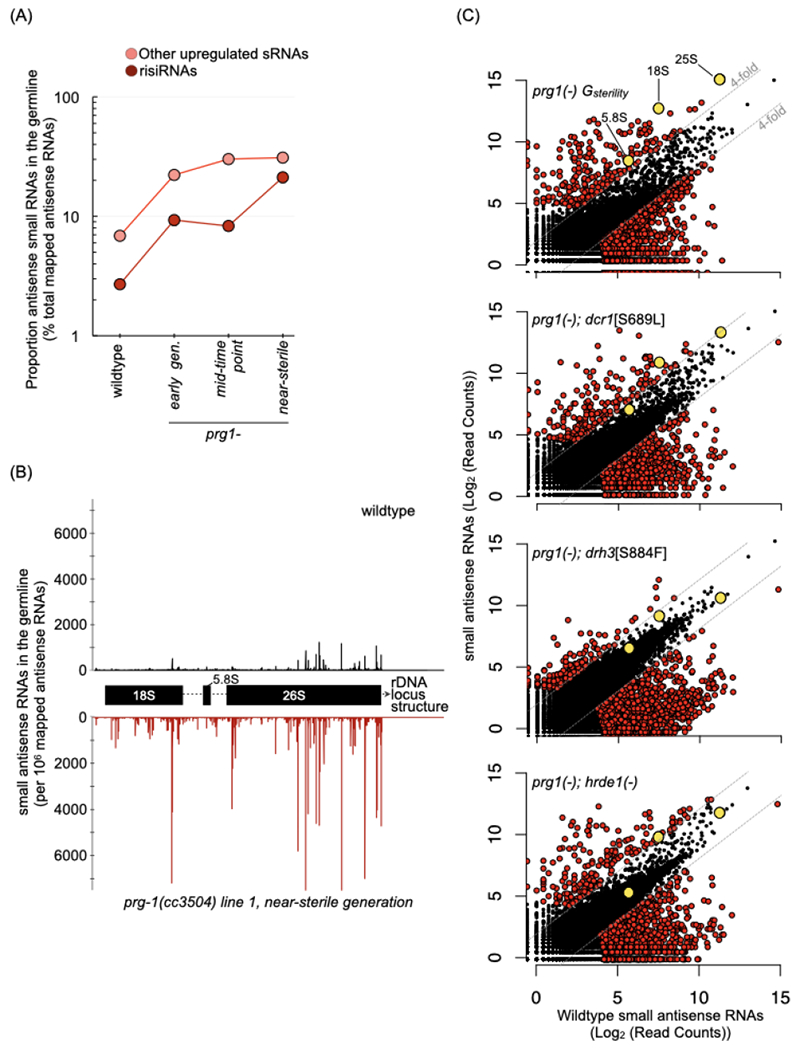
(A) Proportion of risiRNAs and other up-regulated small RNAs (n=215) from near-sterile prg-1(−) in wildtype, and across generations in prg-1(−). Values are percent of total mapped antisense RNAs, and represent the mean of two biological replicates with Gsterility of 12 and 15. Early generation samples are from G1 and G3, mid-time point are from G6 and G8, and near-sterile from G11 and G13, respectively.
See also Figure S5A–B.
(B) Distribution of risiRNAs across the 45S rDNA locus in a wildtype (black) and a near-sterile prg-1(−) (red) line. The 5’ position for each antisense RNA is mapped.
See also Figure S5C.
(C) Scatterplots depicting antisense small RNAs per gene in young adult animals from near-sterile prg-1(−) and three suppressor lines: prg-1(−); dcr-1[S689L], prg-1(−); drh-3[S884F], and prg-1(−); hrde-1(−) relative to wildtype. Counts are per 106 reads mapped to the ce11 genome and represent the mean of two biological replicates. Strains were passaged for 11-14 generations prior to sequencing, to within a generation of full sterility for the prg-1(−) lines passaged concurrently. Grey lines are at 4-fold change relative to wildtype, and red-colored dots indicate genes Colored dots indicate genes meeting the three criteria as in Figure 3A. Grey dots indicate small RNA levels against the three genes in the 45S ribosomal locus (18S, 5.8S and 25S). Number of genes with > 4-fold and < 4-fold small RNAs in the various mutants: prg-1(−) Gsterility= 235 and 2076, prg-1(−);dcr-1[S689L]= 211 and 1003, prg-1(−);drh-3[S884F]= 182 and 1331 and prg-1(−);hrde-1(−)= 199 and 1169.
See also Figure S5D–F.
