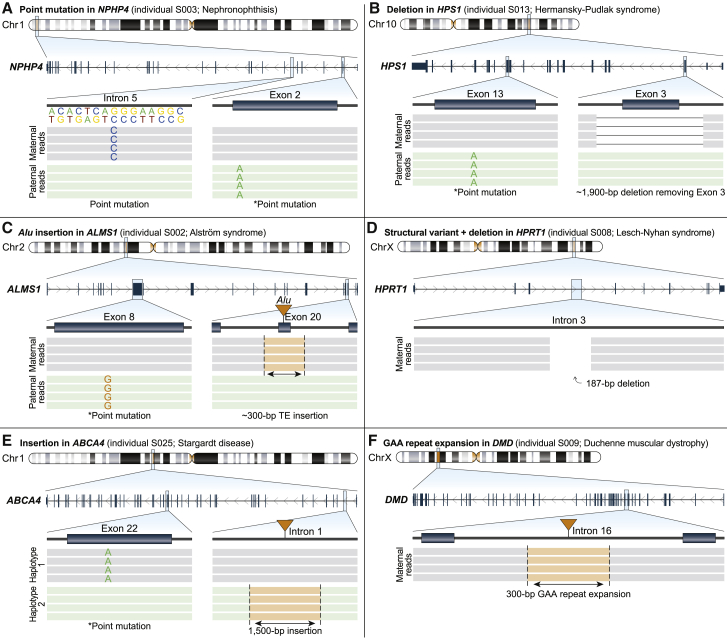
Figure 3.
Targeted long-read sequencing identifies variants not detected by clinical testing
Pathogenic, likely pathogenic, or variants of uncertain significance (VUSs) identified by T-LRS along with variants identified by prior clinical testing (denoted by an asterisk).
(A) T-LRS detected a candidate intronic splice acceptor variant as well as the known paternally inherited stop-gain. Long-read phasing demonstrates that these variants are in trans.
(B) A 1,900 bp deletion within HPS1 removes exon 3; phasing revealed that this variant and the previously known paternally inherited stop-gain occur on different haplotypes. Clinical testing with an exon-level array confirmed the deletion.
(C) T-LRS reveals a previously known paternally inherited stop-gain as well as a novel Alu insertion in exon 20 of ALMS1. Subsequent clinical testing confirmed the Alu was pathogenic and maternally inherited.
(D) A 187 bp deletion and 17 Mbp inversion disrupts HPRT1. Clinical testing confirmed the presence of an inversion.
(E) Insertion of a 1,500 bp composite retrotransposable element is predicted to create multiple splice acceptor and donor sites and represents a candidate second hit. Linkage disequilibrium phasing suggests the variants are on different haplotypes.
(F) Expansion of an AGAA repeat within DMD represents a VUS in an individual with Duchenne muscular dystrophy and a family history lacking a genetic diagnosis.