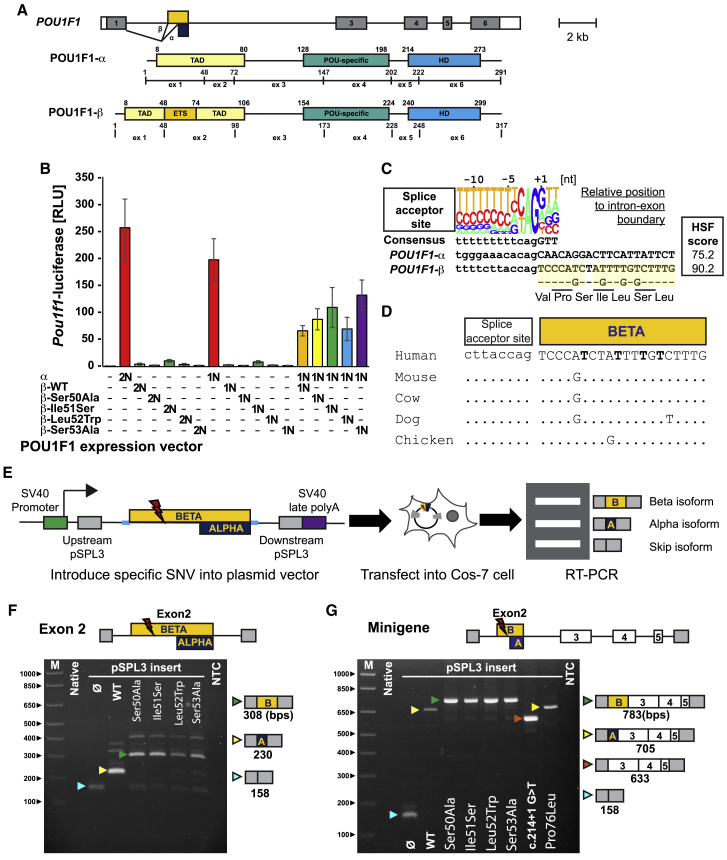
Figure 2.
Variants in the POU1F1 beta coding region suppress the function of the alpha isoform and lead to splicing abnormality
(A) Schematic of the human POU1F1 gene and protein isoforms produced by use of alternate splice acceptors at exon 2. The Pou1f1 beta isoform has an insertion of 26 amino acids located at amino acid 48 in the transactivation domain.
(B) COS-7 cells were transfected with a Pou1f1-luciferase reporter gene and expression vectors for POU1F1 alpha or beta isoforms either singly or together in the ratios indicated (2N and 1N). WT POU1F1 alpha has strong activation at 2N and 1N dosages. WT and variants of the POU1F1 beta isoform have no significant activation over background. A 50:50 mix of alpha and WT beta isoforms exhibited reduced activation. The variant beta isoforms suppress alpha-isoform-mediated activation to a degree similar to WT. The error bars represent standard deviation.
(C) Diagram of the splice acceptor site consensus and the genomic DNA sequence at the boundary between intron 1 and splice sites utilized in exon 2 of POU1F1.41
(D) Evolutionary conservation of the genomic sequence encoding the POU1F1 beta isoform in mammals and chickens.
(E) Exon trapping assay with pSPL3 exon trap vector containing exon 2 of POU1F1 and portions of the flanking introns.
(F) Ethidium bromide-stained gel of exon trap products from cells transfected with the indicated plasmid. Arrowheads indicate the expected products for exon skipping (blue), the alpha isoform (yellow), and the beta isoform (green).
(G) POU1F1 minigenes spanning from intron 1 to 5, with all of the intervening exons, were engineered with the indicated variants and assayed for splicing. WT and p.Pro76Leu POU1F1 splice to produce the alpha isoform, the G>T change in the splice acceptor causes exon skipping (red arrow), and the other variants all splice to produce the POU1F1 beta isoform.