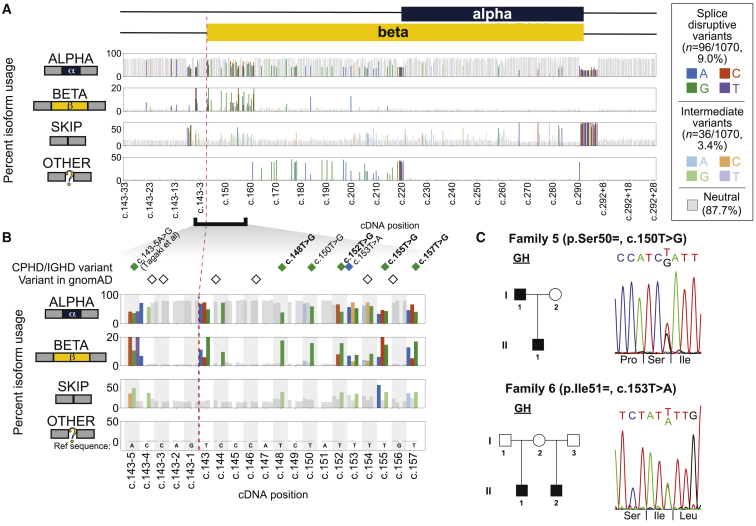
Figure 3.
Splicing effect map in POU1F1 exon 2 and flanking introns and identification of IGHD-affected families with synonymous changes
(A) Percent usage of POU1F1 exon 2 alpha (top panel), beta (second panel), skip (third panel), and other isoforms (bottom panel) by variant position, as measured by massively parallel minigene assay. Gray bars denote splicing-neutral variants, while shaded bars indicate the base pair change of each SDV (dark colors) and intermediate variant (light colors). Cropped intronic regions are shown in Figure S6.
(B) A cluster of SDVs near the beta isoform splice acceptor leads to increased usage of the beta isoform and, in some cases, intermediately increased exon skipping. Diamonds colored by the alternate allele indicate variants in individuals with hypopituitarism, and empty diamonds indicate variants reported in gnomAD. Missense variants’ labels are in bold text.
(C) Families 5 and 6 each had two individuals affected with IGHD and synonymous variants that were splice disruptive. Pedigrees and Sanger sequence confirmation of variants are shown.