Abstract
Bidirectional Encoder Representations from Transformers (BERT) and BERT-based approaches are the current state-of-the-art in many natural language processing (NLP) tasks; however, their application to document classification on long clinical texts is limited. In this work, we introduce four methods to scale BERT, which by default can only handle input sequences up to approximately 400 words long, to perform document classification on clinical texts several thousand words long. We compare these methods against two much simpler architectures – a word-level convolutional neural network and a hierarchical self-attention network – and show that BERT often cannot beat these simpler baselines when classifying MIMIC-III discharge summaries and SEER cancer pathology reports. In our analysis, we show that two key components of BERT – pretraining and WordPiece tokenization – may actually be inhibiting BERT’s performance on clinical text classification tasks where the input document is several thousand words long and where correctly identifying labels may depend more on identifying a few key words or phrases rather than understanding the contextual meaning of sequences of text.
Keywords: BERT, Clinical Text, Deep Learning, Natural Language Processing, Neural Networks, Text Classification
I. Introduction
Document classification is an essential task in clinical natural language processing (NLP). In the clinical setting, labels are often available only at the document level rather than at the individual word level, such as when unstructured clinical notes are linked to structured data from electronic health records (EHRs), and thus document classification is an essential tool in practical automation of clinical workflows. Timely classification of key data elements from clinical documents is extremely important for applications such as precision medicine, population health surveillance, and research and policy. Unfortunately, in the clinical setting, human annotation of EHRs can be extremely time-consuming and expensive due to the technical nature of the content and the expert knowledge required to parse it; thus, effective automated classification of clinical text such as cancer pathology reports and patient notes from hospital stays can make meaningful contributions toward health-related outcomes [1].
Currently, Bidirectional Encoder Representations from Transformers (BERT) [2] and BERT-based approaches achieve state-of-the-art performance in many common tasks within the general NLP community such as question answering, natural language understanding, and text generation. BERT is a computationally expensive deep learning approach that is first pretrained on a very large corpus of unlabelled text on the order of 1 billion or more words – this pretraining step typically takes hundreds to thousands of GPU or TPU hours [3], [4] and allows the model to learn nuanced linguistic patterns that may be useful for downstream tasks. Once pretrained, the model is then fine-tuned on a specific task of interest. To limit the vocabulary size and generalize better to new words outside the training vocabulary, BERT utilizes subword-level WordPiece tokens rather that word-level tokens as input.
Adapting BERT to the task of clinical document classification poses non-trivial challenges. First, most BERT-based implementations have a maximum input length of 512 WordPiece tokens, which is roughly equal to 400 words. Unfortunately, clinical documents can very easily exceed this limit – the average discharge summary in the MIMIC-III dataset is approximately 2000 word tokens [5]. Second, to maximize performance, BERT-based models must be pretrained on a text corpus that is from a similar domain as the downstream application task. Therefore, clinical practitioners who wish to apply BERT-based models but do not have access to the compute or data necessary to pretrain their own models must rely on downloading existing pretrained models such as BioBERT [6] or BlueBERT [7]. Some recent work, such as the Reformer [8] and LongFormer [9] models, adapt BERT for longer input texts; however, at the time of this study, there exist no publicly available pretrained weights in the biomedical and/or clinical domain for these models. For this reason, we utilize BlueBERT, which is the original BERT model pretrained on sentences from Pubmed abstracts and MIMIC-III clinical notes, as the main model for this work.
In this work, we test different methods to adapt BlueBERT for text classification on long clinical documents – these methods consist of splitting long documents into smaller chunks, processing the chunks individually, and then combining the outputs using max pooling or attention-based methods. We apply these methods to both the single-label and multilabel classification settings. We compare the performance of BlueBERT against two strong baselines – a shallow, word-level convolutional neural network (CNN) [10] and a hierarchical self-attention network (HiSAN) [11], both of which have nearly two orders of magnitude fewer learnable parameters than BERT. We show that BERT actually achieves similar performance to the CNN and underperforms the HiSAN on many of the clinical document classification tasks that we test on. Our contributions are as follows:
We compare the effectiveness of different ways to adapt BERT for document classification on long clinical texts up to several thousand words in length
We evaluate the effectiveness of BERT on clinical single-label and multilabel document classification against two other strong baselines – the CNN and the HiSAN
We show that a much simpler deep learning model, the HiSAN, can obtain similar or better performance compared to BERT on many of our clinical document classification tasks
To better understand the weaknesses of BERT in our tasks, we analyze the attention weights within the HiSAN and BERT to understand how each model identifies keywords and show that using WordPiece subword tokens may be more difficult than using word-level tokens
II. Related Work
While BERT and BERT-based models have achieved state-of-the-art performance across a wide range of various NLP tasks including question answering [12], [13], information extraction [14], [15], and summarization [16], [17], their applications to long document classification tasks have been extremely limited. To our knowledge, there exists only one previous in-depth study on strategies to adapt BERT for long document classification: Sun et al [18] explore different techniques for using BERT to classify moderate-length documents from IMDb reviews, Yelp reviews, Sogou News, and other similar datasets. The study finds that the best overall classification accuracy is achieved by using only the first 128 and the last 382 tokens in each document as the input into BERT and dropping all intermediate content. While other works [19–21] have applied BERT to text classification related tasks, none explore the problem of long-document inputs that are longer than BERT’s default max input length of 512 WordPiece tokens.
There are several reasons that the findings from [18] may not hold in the clinical document domain. First, most of the datasets tested in [18] are moderate in length – for example, only 12.69% of the documents in IMDb exceed 512 tokens in length, 4.60% in Yelp, and 46.23% in Sogou, and even in Sogou the average length is only 737 tokens. Thus, it is uncertain how BERT will perform on datasets such as MIMIC-III where the average discharge summary is over 2000 tokens long. Second, in clinical documents classification tasks, the presence of a specific label may be indicated by only a short phrase that appears only once in the entire document; therefore, using only the first 128 and the last 382 tokens may be more detrimental than in a task such as news classification or sentiment analysis, where context clues may be scattered throughout the document.
In the clinical and biomedical domain, BERT has been applied to various tasks that do not include document-level classification. BioBERT [6], which is pretrained on PubMed abstracts of PMC full-text articles, showed superior performance on biomedical named entity recognition, relation extraction, and question answering tasks. ClinicalBERT [22], which starts with BioBert and then further pretrains on MIMIC-III clinical notes, showed superior performance on clinical natural language inference tasks. BlueBERT [7], pretrained on PubMed abstracts and MIMIC-III clinical notes, achieved superior performance on biomedical and clinical sentence similarity, named entity recognition, relation extraction, and short document classification tasks. Two common characteristics of all these tasks are (1) input length is less than or equal to 512 WordPiece tokens and (2) understanding sequences of words in context is generally critical to the task. In [23], authors pretrained their own BERT model on Italian clinical text, applied it to Italian pathology report classification, and found that BERT underperforms more simple architectures, but the study focused on short inputs less than 512 WordPiece tokens in length. BERT has yet to be thoroughly tested under settings where the input document is several thousand words long and where correctly identifying labels may depend more on identifying a few key words or phrases rather than understanding the contextual meaning of sequences of text.
The current state-of-the-art approaches for clinical document classification are generally models that pre-date contextual word embedding-based approaches such as BERT. Clinical NLP approaches often lag behind those used in the general NLP community partly due to the legal challenges of releasing open research datasets to promote the development of new approaches [24], [25]. Recent approaches for clinical document classification include rule-based methods [26], [27], traditional machine learning [28], [29], convolutional neural networks (CNNs) [30], [31], recurrent neural networks (RNNs) [32], [33], and self-attention networks [11].
In this work, we compare different strategies to adapt BERT to long documents against existing strong baselines using discharge summaries from the MIMIC-III dataset and cancer pathology reports obtained from Louisiana Tumor Registry, Kentucky Cancer Registry, Utah Cancer Registry, and New Jersey State Cancer Registry. There are three multilabel classification tasks for MIMIC-III – diagnostic codes, diagnostic categories, and procedure codes – and six single-label classification tasks for the cancer pathology reports – identifying cancer site, subsite, laterality, behavior, histology, and grade.
III. Materials and Methods
A. BERT for Document Classification
In this work, we begin with the assumption that end-users wish to apply BERT to their document classification tasks but lack the computational resources and/or training data on the order of 1B+ words required to pretrain BERT from scratch; thus, users must start from an available pretrained model. Because we are working with clinical text documents, we utilize BlueBERT [7], which is the BERT model pretrained on PubMed abstracts and MIMIC-III clinical notes. As the architecture of BERT has been widely described and explored in existing literature, we refer the reader to those studies [2], [34], [35] to learn about the base architecture of the BERT model.
Because the self-attention mechanism used in BERT has memory requirements that scale quadratically based off the sequence length, the original BERT model was primarily designed to handle sentence-length and paragraph-length inputs and has a maximum input length of 512 WordPiece tokens, or roughly 400 word tokens. As a result, subsequent BERT-based models pretrained on different corpora, including BioBERT, Clinical BERT, and BlueBert, all share this same limitation on input length. To adapt BERT for long document classification, we explore the following strategies (illustrated in Figure 1):
Fig. 1:
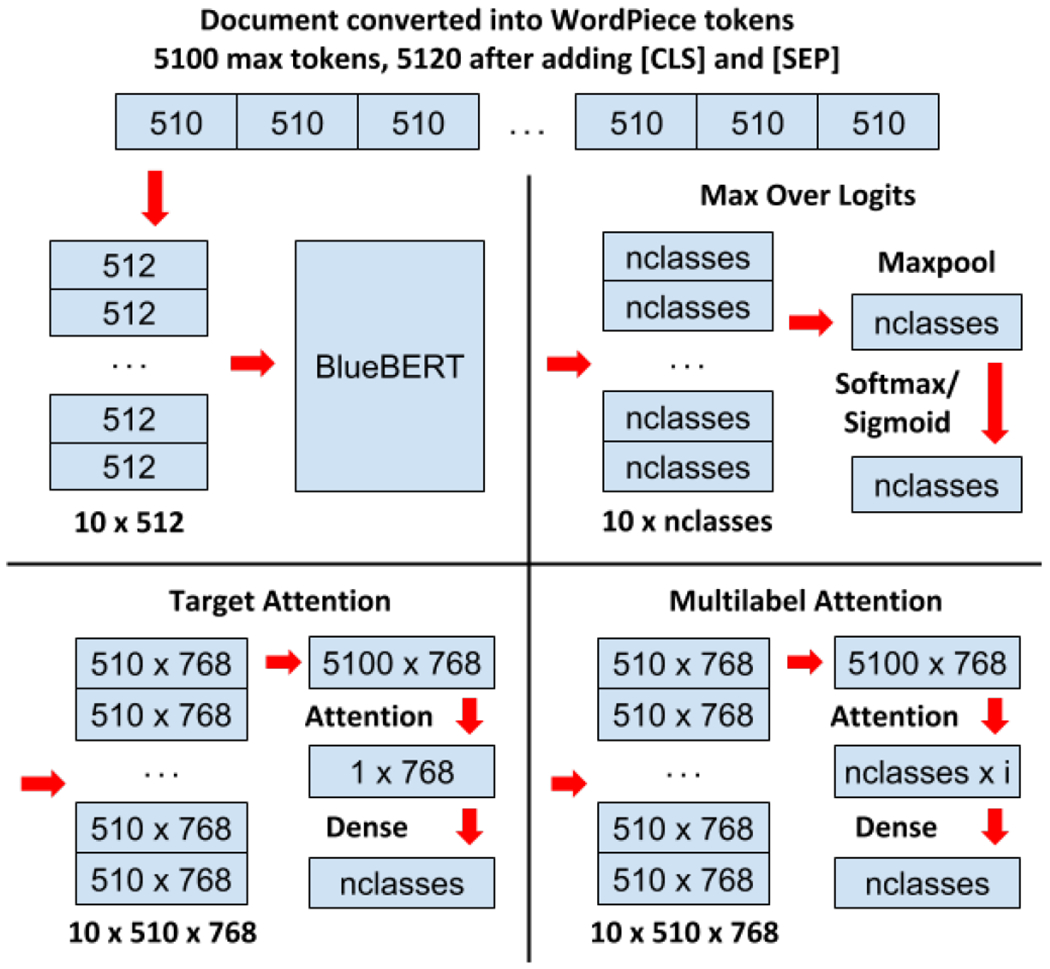
Process for splitting long documents into smaller chunks to feed into BERT and methods for combining the resulting BERT outputs from each chunk into a single classification decision.
1). First 510 WordPiece Tokens Only:
For any input document, we convert the document into WordPiece tokens and use only the first 510 tokens. As standard practice for BERT-based models [2], each token sequence is prepended by the [CLS] token (used for classification) and appended by the [SEP] token (marks the end of an input sequence for one or more input sequences), making a total of 512 tokens, the maximum input length for BERT. As BERT is already preconfigured for a wide range of tasks including sequence classification [2], we use the standard sequence classification setup where the output of the [CLS] token is then fed into an intermediate dense layer and a final classification layer. For single-label classification, the output logits from the classification layer are fed into a softmax activation, whereas for multilabel classification, the logits are fed into a sigmoid activation.
We note that this strategy may discard a significant portion of content for each document that may be useful for classification; therefore, we expect that this strategy may perform poorly due to information loss. However, we include this strategy as it is useful to establish a baseline.
2). Max Pool Over Logits:
In order to capture the content from the entire document, we utilize a hierarchical approach in which we split long documents into smaller chunks and then process each chunk individually using BERT. After converting an input document into WordPiece tokens, we split the document into k segments of 510 tokens each. Each segment is prepended by the [CLS] token and appended by the [SEP] token so that it is 512 in length. We then utilize the standard BERT classification setup on each of the k segments, wherein the first [CLS] token in each segment is passed to an intermediate dense layer and a final classification layer. This generates k logit vectors, one for each segment.
Prior to the softmax or sigmoid activation function, we apply a max pool operation across all k logits to reduce them into a single logit vector – this max pooled logit vector represents the maximum logit value for each possible class across each of the k segments. For single-label classification, this final max pooled logit vector is passed to a softmax activation to predict class probabilities, and for multilabel classification it is passed to a sigmoid activation.
We note that max pooling is performed on the logit vector because the size of the logit vector is always equal to the number of possible classes and a higher logit value for a given class will always indicate that particular class is more likely to be present. This cannot be said about any other intermediate representation generated by BERT, where a large negative value may be just as important as a large positive value in identifying the presence of a particular class. Thus, applying max pool to the logit vector minimizes unintentional information loss.
3). Target Attention:
Similar to max pool over logits, we split the document into k segments of 510 WordPiece tokens each. Each segment is prepended by the [CLS] token and appended by the [SEP] token so that it is 512 in length. We then utilize the BERT model without the classification setup such that for each of the k segments, we simply generate 512 new contextual token embeddings. From this, we drop the first [CLS] and last [SEP] token embeddings from each of the k sequences, then concatenate the k embedding sequences to form , where l is the total length of the document and d is the embedding dimension configured within BERT (768 in our case).
Next, we utilize an attention mechanism to identify the token embeddings within E that are most relevant to the target task. To do this, we utilize a modified version of scaled dot product attention [36], which is shown in Equation 1:
| (1) |
where and are learnable weight matrices and and are learnable bias vectors. and are simple linear transformations of E. Finally, is a randomly initialized vector that is learned through training – this vector represents the information to look for given the current task.
Essentially, our target attention operation compares each token embedding in E to the target vector T to identify the embeddings most relevant to the current task. The output of our target attention mechanism is , the final document embedding used for classification, which is effectively a weighted average of the most important embeddings from E. We pass D to a final dense classification layer; as the previous strategies, the output logits from the classification layer are fed into a softmax activation for single-label classification and a sigmoid activation for multilabel classification.
4). Multilabel Attention:
In the multilabel classification setting, we expand our target attention mechanism so that we use a separate parallel attention mechanism for each possible label. This increases the expressivity of the attention mechanism so that the same attention target vector does not need to capture information for hundreds or thousands of possible labels.
Once again, we split the document into k segments of 510 WordPiece tokens each. Each segment is prepended by the [CLS] token and appended by the [SEP] token so that it is 512 in length. We use the same procedure from target attention to generate , which represents the contextual embeddings generated by BERT for the all tokens in the document. We then pass E to a modified version of scaled dot product attention, shown in Equation 2:
| (2) |
where and are learnable weight matrices and and are learnable bias vectors. and are simple linear transformations of E. Unlike in target attention where the embedding dimension of K and V are set to d, the same as E, in multilabel attention they are reduced to an intermediate dimension i as we found this reduces overfitting. is a randomly initialized matrix that is learned through training, where c is the number of possible classes – each row of this matrix represents the most important information for one class.
While in target attention each embedding in E is compared to a single target vector to determine its relevance, in multilabel attention each embedding in E is simultaneously compared to a different vector for each possible class to determine its relevance for that class. The output of multilabel attention is a matrix , which we pass to a dense layer with weights and bias to generate the logits. Because we only utilize multilabel attention in the multilabel classification setting, we pass the output logits to a sigmoid activation to obtain the final class probabilities.
B. Baseline Models
1). Convolutional Neural Network:
Our first strong baseline is a shallow word-level CNN based off [37]. Although a relatively simple architecture that was originally developed in 2014, it is still widely used for biomedical and clinical text classification and has shown strong performance across a variety of tasks [10], [38–40]. For our CNN implementation, we represent each document using word level embeddings, which are passed to three parallel 1D convolution layers; these examine three, four, and five consecutive words at a time to identify n-grams relevant to the given task. The outputs from the three convolution layers are concatenated and passed to a max pool operation that generates a fixed-length document vector composed of the most important n-grams in the document. This document vector is passed to a final dense classification layer that uses softmax for single-label classification and sigmoid for multilabel classification.
In multilabel classification settings, we also test a multilabel variant of the CNN, which we refer to as CNN-multilabel (CNN-ML). In this variant, after the outputs from the three convolution layers are concatenated, instead of using a max pool operation, we feed the output to the same multilabel attention setup that we use for BERT.
2). Hierarchical Self-Attention Network:
Our second strong baseline is the HiSAN network [11], which to our knowledge is the current state-of-the-art in classifying cancer pathology reports. Like BERT, this architecture is also based off self-attention operations, but it is far simpler and has approximately 100x fewer learnable parameters. We use the exact same implementation as [11] – first, each document is represented using word level embeddings and then broken into chunks of ten words each. The HiSAN’s lower hierarchy uses a series of attention-based operations to generate a fixed-length vector representation for each ten-word chunk. These representations are then passed to the HiSAN’s upper hierarchy, which uses another series of attention-based operations to generate a fixed-length vector representation of the entire document. This document vector is passed to a final dense classification layer that uses softmax for single-label classification and sigmoid for multilabel classification.
Like with the CNN, in the multilabel classification setting we test a multilabel variant of the HiSAN, which we refer to as HiSAN-multilabel (HiSAN-ML). In this variant, we replace target attention mechanism in the HiSAN’s upper hierarchy with the same multilabel attention setup that we use for BERT.
C. Datasets
1). MIMIC-III Discharge Summaries:
The MIMIC-III dataset consists of unstructured clinical notes as well as structured tables related to 49,785 distinct hospital admissions of 38,597 unique adult patients who stayed in the intensive care unit at Beth Israel Deaconess Medical Center between 2001 and 2012 [5]. Each unique admission is annotated by human experts with a set of ICD-9 codes that describe the diagnoses and procedures that occurred during that particular stay. Each unique admission is also associated with a discharge summary which summarizes the information from the stay in a single document. For this study, we utilize the discharge summaries for three multilabel classification tasks – (1) predict the set of 5-character diagnoses codes (DX 5-Char) associated with each discharge summary, (2) predict the set of unique 3-character (DX 3-Char) diagnoses categories associated with each discharge summary, which consists of the first three characters of the full 5-character diagnosis code, and (3) predict the set of procedure codes associated with each discharge summary.
We note that some admissions have one or more addenda in addition to the discharge summary; in these situations we concatenate the information from the addenda to the discharge summary. Following [31], we perform train/val/test splitting based off unique patient IDs so that the same patient does not appear in multiple splits. Statistics regarding this dataset are available in Table I.
TABLE I:
Dataset descriptions for each task. We note that document lengths are measured using generic word tokens rather than BERT’s WordPiece tokens. Converting to WordPiece tokens results in approximately 25% more tokens.
| Train Docs | Val Docs | Test Docs | Unique Labels | Avg Labels Per Doc | Avg Words Per Doc | Std Dev Words Per Doc | |
|---|---|---|---|---|---|---|---|
| MIMIC-III: DX 5-Char | 42262 | 5223 | 5241 | 6919 | 11.7 | 2061 | 992 |
| MIMIC-III: DX 3-Char | 42262 | 5223 | 5241 | 942 | 10.8 | 2061 | 992 |
| MIMIC-III: Procedure | 42262 | 5223 | 5241 | 1990 | 4.5 | 2061 | 992 |
| Pathology Reports: Site | 144754 | 25545 | 30053 | 70 | 1 | 622 | 465 |
| Pathology Reports: Subsite | 144754 | 25545 | 30053 | 325 | 1 | 622 | 465 |
| Pathology Reports: Laterality | 144754 | 25545 | 30053 | 7 | 1 | 622 | 465 |
| Pathology Reports: Histology | 144754 | 25545 | 30053 | 578 | 1 | 622 | 465 |
| Pathology Reports: Behavior | 144754 | 25545 | 30053 | 4 | 1 | 622 | 465 |
| Pathology Reports: Grade | 144754 | 25545 | 30053 | 9 | 1 | 622 | 465 |
2). SEER Cancer Pathology Reports:
The National Cancer Institute (NCI) Surveillance, Epidemiology, and End Results (SEER) program works with cancer registries across the United States to collect and maintain cancer data in order to support national cancer surveillance. We obtained 1,201,432 cancer pathology reports from the Louisiana, Kentucky, New Jersey, and Utah SEER cancer registries. Each cancer pathology report is associated with a unique tumor ID; one or more cancer pathology reports may be associated with the same tumor ID. For each tumor ID, certified tumor registrars (CTRs) manually assigned ground truth labels for key data elements – including cancer site, subsite, laterality, behavior, histology, and grade; for a given tumor ID, labels were assigned based off all data available for that tumor ID. Because our ground truth labels are at the tumor level rather than the report level, there are cases where tumor IDs associated with multiple pathology reports have labels which do not match the content within one or more of the individual pathology reports. Therefore, in this study we only utilize tumor IDs associated with a single pathology report, yielding a total of 200,352 pathology reports. We utilize this dataset to perform six single-label document classification tasks, one for each manually annotated data element. Statistics regarding this dataset are available in Table I.
IV. Experiments
A. Evaluation Metrics
For multilabel classification tasks on the MIMIC-III dataset, we follow established metrics from previous work [30–32] and measure performance using precision, recall, and F1 score, where each possible text-code pair is treated as an independent prediction:
| (3) |
| (4) |
| (5) |
Similarly, for single-label classification tasks on the cancer pathology reports, we follow established metrics from previous work [10], [11], [33], [41] and measure performance using classification accuracy and macro F1 score, in which the F1 score is calculated for each possible class label and then averaged across all class labels:
| (6) |
where Ci represents the subset of training samples belonging to class i, and |C| is the total number of possible classes. Because of the extreme class imbalance inherent in the cancer pathology report dataset, macro F1 score better captures model performance on minority classes.
For all metrics, we bootstrap samples from our test set using a procedure described in Appendix A to generate 95% confidence intervals. Since computation speed may also be a consideration in some applications, we report the average inference time for 1000 documents for each method on the MIMIC-III dataset utilizing a single Tesla V100 GPU.
B. Dataset Cleaning
For both datasets, we lowercase all text, clean hex and unicode symbols, replace decimal values and integers larger than 100, and clean up any deidentification tokens; a more detailed description is available in Appendix B. For BERT-based approaches, we utilize the HuggingFace BERT tokenizer1 with the vocabulary associated with the pretrained BlueBERT model2. For the CNN and HiSAN, we train 300-dimensional word2vec embeddings on each dataset, replacing unique words appearing in fewer than five documents in each dataset with an “unknown_word” token.
C. Hyperparameter Optimization
For all BERT-based approaches, we start from pretrained weights from BlueBERT Base2 and implement all models using the Huggingface library1. For the max pool over logits, target attention, and multilabel attention methods, we limit the number of segments per document k to a maximum of 10; we note that k is not a tuned hyperparameter but instead determined based off the average length of our documents and the memory capacity of our Tesla V100 GPUs.
Hyperparameters for all approaches are optimized using the validation set of each dataset. Due to the high computational cost of some of our models, we use a hill-climbing strategy in which we change a single hyperparameter at a time and then retrain until model performance stops improving. We choose the set of hyperparameters with the overall highest performance across all tasks (average F1 for MIMIC and average accuracy for pathology reports). We list the range of hyperparameters explored as well as the optimal hyperparameters in Table II.
TABLE II:
Hyperparameters explored for each model.
|
BERT
| |
| Multilabel Attention Dim | 100, 200, 300*^, 400, 500 |
|
| |
| Batch Size | 8, 16*^, 32, 64 |
|
| |
| Adam Learning Rate | 5E-6, 1E-5, 2E-5*^, 5E-5 |
|
| |
|
CNN
| |
| Filter Size | 100, 300^, 500, 1000* |
|
| |
| Dropout % | 0, 10, 15*, 25, 50^ |
|
| |
| Multilabel Attention Dim | 100, 200, 300, 400*^, 500 |
|
| |
| Batch Size | 32, 64, 128*^, 256 |
|
| |
| Adam Learning Rate | 5E-5, 1E-4*^, 2E-4, 5E-4 |
|
| |
|
HiSAN
| |
| Attention Size | 400, 512^, 768*, 1024 |
|
| |
| Attention Heads | 4, 8*^, 16 |
|
| |
| Dropout % | 0, 10*^, 15, 25, 50 |
|
| |
| Multilabel Attention Dim | 100, 200, 300, 400*^, 500 |
|
| |
| Batch Size | 32, 64, 128*^, 256 |
|
| |
| Adam Learning Rate | 5E-5, 1E-4*^, 2E-4, 5E-4 |
Optimal hyperparameters are marked with a for the MIMIC III tasks
for the pathology report tasks.
D. Results
Figure 2 shows the results of our experiments on the MIMIC-II dataset dataset. First, we examine the performance of each model when limited to only the first 510 WordPiece tokens. We note that for models such as the CNN and HiSAN that use word token inputs, we convert the first 510 WordPiece tokens back into word tokens which results in approximately 400 word tokens for each document. We use this first set of results to address two key questions: (1) how well does each method perform when using only the first 510 WordPiece tokens compared to the full document and (2) how well does BlueBERT compare to our strong baselines when adaptive methods to fit longer documents isn’t a performance factor?
Fig. 2:
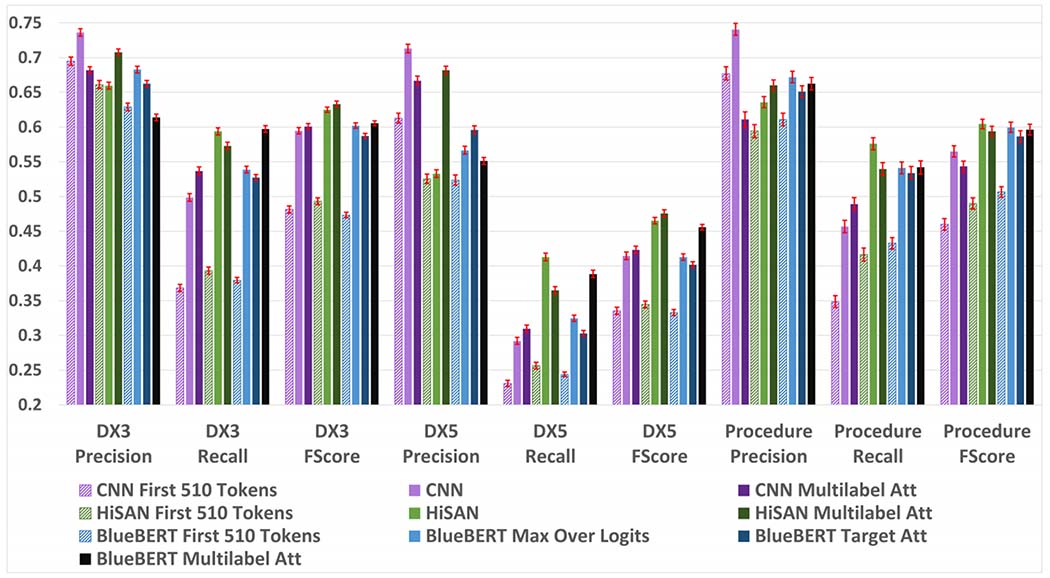
Precision, recall, and F1 scores for each model on the MIMIC-III dataset. 95% confidence intervals are shown in red and calculated using a bootstrapping procedure detailed in Appendix A.
Our results in Figure 2 suggest that even if we limit all models to short text segments that fit within BERT’s default 512 WordPiece input limit, BERT does not outperform our much simpler baselines in two of the three tasks. The CNN model consistently achieves the best precision scores by a wide margin on all tasks. We expect that this because the CNN is designed to memorize the 3-, 4-, and 5-gram word combinations associated with each label as opposed to learning more complex sequential patterns; this limits the ability of the CNN to generalize beyond the n-gram patterns it knows, but makes it very precise when it does encounter a previously seen n-gram. The HiSAN and BERT models can both learn more complex patterns than the CNN and achieve better recall than the CNN on all tasks at the cost of precision. The HiSAN model achieves the best recall and F1 scores on the diagnosis category and full code tasks, whereas BERT achieves the best recall and F1 score on the procedure task.
Second, we examine the performance of each model using full documents from the MIMIC-III dataset. We notice that compared to using only the first 510 WordPiece tokens, using the full document results in significantly improved performance across all metrics. This makes intuitive sense, as on average, the first 510 WordPiece tokens captures approximately only the first 25% of each document and critical information may be located in the remainder of the document.
When using full documents on the MIMIC-III dataset tasks, our BERT-based approaches do not significantly outperform our much simpler baselines on any tasks. Once again, the CNN model consistently achieves the best precision scores by a wide margin on all tasks. The HiSAN-based approaches achieve the best recall and F1 scores on most tasks; while the BERT multilabel attention approach achieves the best recall score on the diagnostic category task, it is not significantly better than that of the HiSAN model.
When comparing the different methods for adapting BERT to longer text documents, the max over logits method consistently outperforms the target attention method in all tasks and metrics except for precision score in the diagnostic code task. Interestingly, using multilabel attention has mixed effects based on both task and model. For the CNN and BERT models, multilabel attention increases recall at the cost of precision, whereas for the HiSAN model it increases precision at the cost of recall. For all models, multilabel attention appears to help most in the diagnostic category and code tasks while having mixed results in the procedure task.
Figure 3 shows the results of our experiments on the cancer pathology reports dataset. After taking into account confidence intervals, BERT does not achieve statistically better accuracy or macro F1 scores than the HiSAN on any of the six tasks. Similar to our results from the MIMIC-III dataset, the max over logits approach almost always performs better than the target attention approach on all tasks and metrics.
Fig. 3:
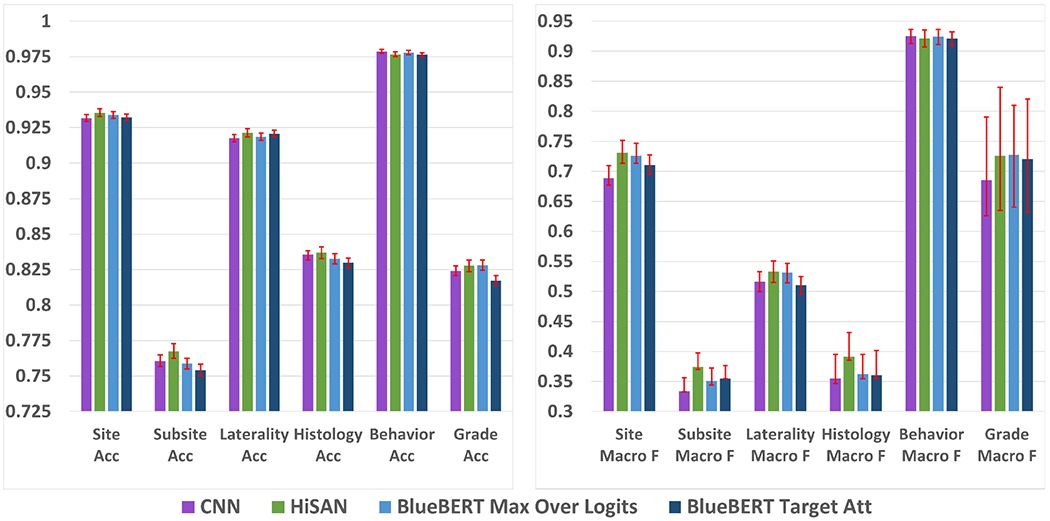
Accuracy (left) and macro F1 scores (right) for each model on the cancer pathology report dataset. 95% confidence intervals are shown in red and calculated using a bootstrapping procedure detailed in Appendix A.
Finally, Table III shows the average time in seconds to predict on 1000 full documents from the MIMIC-III dataset. We see that the BERT-based approaches are almost an order of magnitude slower than the base CNN and the HiSAN-based models. While inference time may not be the most critical factor for institutions that only need to perform a single prediction pass on their data, it may be important for institutions that have millions of documents or need to regularly retrain their models on incoming data.
TABLE III:
Average time (in seconds) to predict on 1000 full documents from the MIMIC-III dataset. All timing is performed on a DGX machine using a single V100 GPU.
| MIMIC-III DX 3 | MIMIC-III DX 5 | MIMIC-III Procedure | |
|---|---|---|---|
| CNN | 8.4413 | 8.4886 | 8.4537 |
|
| |||
| CNN Multilabel | 18.2624 | 22.5554 | 19.0566 |
|
| |||
| HiSAN | 8.0586 | 8.1274 | 8.1222 |
|
| |||
| HiSAN Multilabel | 8.4029 | 8.6632 | 8.4700 |
|
| |||
| BlueBERT Base | 75.1377 | 75.2986 | 75.2502 |
| Max Over Logits | |||
|
| |||
| BlueBERT Base | 75.8992 | 76.0667 | 75.9519 |
| Target Attention | |||
|
| |||
| BlueBERT Base | 75.8698 | 77.6511 | 76.1592 |
| Multilabel Attention | |||
V. Discussion
Our experiments show that BERT generally does not achieve the best performance on our clinical text classification tasks compared to the much simpler CNN and HiSAN models. In this section, we provide evidence for two potential explanations for the weak performance of BERT – attention dilution and difficulty of subword tokens.
First, one of the key components of BERT’s previous success is the masked-language modelling pretraining process, in which the BERT model may learn subtle and complex word relationships between all possible words in a large unlabelled text corpus. However, in clinical text classification tasks on documents in which only very few words contribute toward a specific label, most of these subtle word relationships may not be necessary or even relevant to the task at hand. Therefore, BERT’s attention may actually be diluted away from the keywords most critical to the task.
To demonstrate this phenomena, we generated three different types of attention visualizations. First, we multiplied the attention weights through both hierarchies of the HiSAN to show exactly which words the HiSAN focuses on in each document (first 510 WordPiece tokens only). Second, using our fine-tuned BlueBERT model (first 510 WordPiece tokens only), we visualized the attention weights from the very final layer that are associated with the [CLS] token used for classification; these weights represent the most important subword tokens after they have already incorporated contextual information from other subword tokens based off the 12 self-attention layers of the main BERT model. Third, using our fine-tuned BlueBERT model (first 510 WordPiece tokens only), we started from the attention weights from the very final layer that are associated with the [CLS] token and multiplied these attention weights through all 12 self-attention layers of the BERT model; these weights represent the most important subword tokens accounting for all the inter-word relationships captured during pretraining and fine-tuning. We provide an example of these visualizations in Appendix C.
After examining these attention weights over a large number of documents, we noticed that in general, (1) the attention weights in the final layer of BERT are more spread out and less focused on specific biomedical keywords than the attention weights from the HiSAN, and (2) the attention weights when accounting for all layers of BERT are even more diluted than those from the final layer of BERT. While there is usually some overlap in the attention weights of the HiSAN and BERT, we found that in a notable number of cases BERT places emphasis on less relevant tokens such as punctuation and [SEP]. These visualizations suggest that BERT’s attention is diverted toward word relationships learned during pretraining as opposed to the specific keywords relevant to the downstream classification task.
Second, while the HiSAN and CNN models utilize word-level tokens as input, BERT uses a WordPiece tokenizer that splits each word into one or more subword tokens. Whereas with word level tokens, the HiSAN and CNN can directly memorize keywords or keyphrases important to each label, there is an added layer of complexity with WordPiece tokens in that important keywords may be broken into multiple wordpiece tokens. Thus, critical keywords or keyphrases will always be longer when represented as WordPiece tokens compared to word-level embeddings, thereby increasing the complexity of the token combinations that a model must learn to recognize a particular label.
To test this hypothesis, we retrained the CNN and HiSAN models on the MIMIC-III dataset using the first 512 subword tokens generated by the final layer of the BlueBERT model (without any fine-tuning on the MIMIC-III dataset) instead of using word-level Word2Vec embeddings. Our results are shown in Figure 4. We see that compared to using word-level tokens as input, both the CNN and HiSAN trained on subword token inputs perform worse in overall F1 score across all three tasks. These results suggest that overall, it may be more difficult to use subword-level tokens for our MIMIC-III classification tasks than it is to use word-level tokens.
Fig. 4:
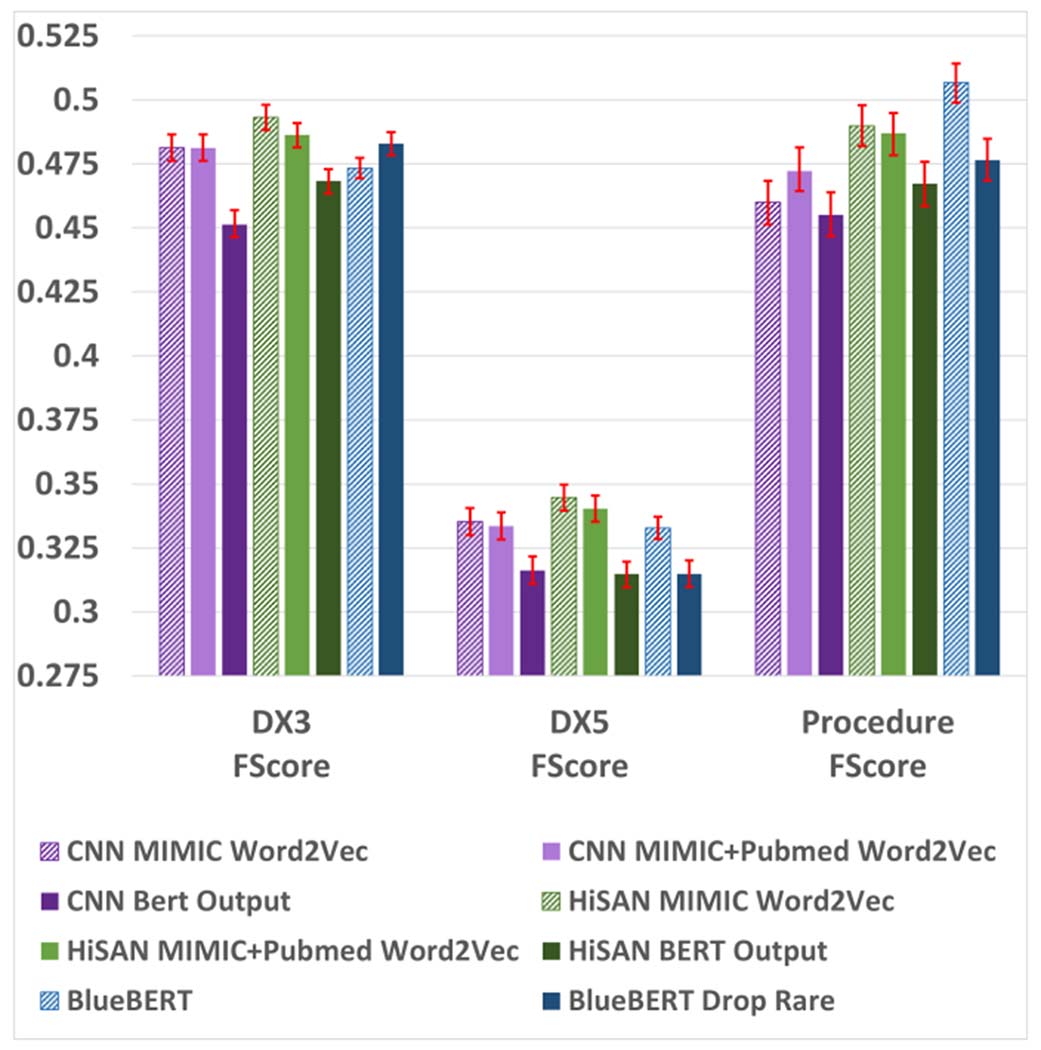
F1 scores for alternative vocabulary/tokenization setups on the MIMIC-III dataset (first 510 WordPiece tokens only). 95% confidence intervals are shown in red and calculated using a bootstrapping procedure detailed in Appendix A.
Finally, we examined differences in the vocabulary and tokenization setups between BERT and our baseline models as a source of performance discrepancy. In our main experiments, we used word embeddings trained directly on the target corpus for our baseline models, eliminating word tokens appearing fewer than five times, whereas for BlueBERT we tokenized using the associated WordPiece vocabulary pretrained on Pubmed abstracts and MIMIC-III. Therefore, we tested (1) the performance of our baseline CNN and HiSAN when utilizing publicly available word embeddings pretrained on Pubmed and MIMIC-III [42], and (2) the performance of BlueBERT when eliminating rare words appearing fewer than five times in the target corpus (mirroring the original tokenization process for the CNN and HiSAN). The results of these experiments on the MIMIC-III dataset (first 510 Wordpiece tokens) are shown in Figure 4. Using pretrained word vectors generally results in slightly worse F1 scores for the CNN and HiSAN, but both models still outperformed BERT in the same two out of three tasks. Eliminating rare tokens reduced BERT’s F1 score in two of the three tasks, and in all three tasks the F1 score was worse than that of the HiSAN. Ultimately, we see that BlueBERT still does not consistently outperform the CNN and HiSAN baselines under any of these alternative tokenization setups.
VI. Conclusion
In this work, we compared four methods for adapting BERT, which by default can only take inputs of up to 510 WordPiece subword tokens, to sequence classification on long clinical texts up to several subword tokens in length; these methods include using only the first 510 WordPiece tokens, hierarchical max pool over logits, hierarchical target attention, and hierarchical multilabel attention. We compare these methods against two strong baselines, the CNN and the HiSAN. We evaluted these models on two datasets. The MIMIC-III clinical notes dataset has three multilabel classification tasks: diagnosis codes, diagnosis categories, and procedure codes; and the cancer pathology reports dataset has six single label classification tasks: site, subsite, laterality, histology, behavior, and grade.
Our results showed that on most datasets and tasks, the BERT-based methods performed equal to or worse than the simpler HiSAN baseline, and in some cases BERT performed equal to or worse than even the CNN. On the MIMIC-III dataset, when all models and baselines were limited to the first 510 WordPiece tokens of each document only, BERT outperformed in only the recall metric for the procedure code task. Once we utilized full length documents, BERT outperformed on only the recall metric for the diagnostic category task. On the cancer pathology report dataset, BERT was not statistically better than the HiSAN on any of the six tasks. Within the four different methods for adapting BERT to classification on long texts, hierarchical multilabel attention had the overall strongest performance on multilabel classification and hierarchical max pool over logits had the overall strongest performance on single label classification.
In our analysis, we presented evidence for two possible reasons why BERT underperforms in clinical text classification tasks. First, our tasks generally have a low signal-to-noise ratio, in that the presence of a few keywords may be enough to indicate a particular label. In BERT’s pretraining process, BERT learns complex and nuanced relations between all words in the pretraining corpus; however, many of these relationships may be irrelevant for the classification task and may actually divert attention away from the critical keywords. Second, BERT’s WordPiece tokenizer breaks each word token into one or more subword tokens. This increases the complexity of the classification task, as now the model must learn to associate a larger number of subword tokens to each label compared to a lower number of word-level tokens.
Our results suggest that a pretrained BERT model such as BioBERT or BlueBERT may not be the best choice for clinical text classification tasks, and a simple CNN or HiSAN model may achieve comparable or better accuracy/F1. However, recent work may address some of BERT’s limitations that we illustrated. For example, [43] utilizes a novel pretraining technique that forces BERT to focus on learning knowledge about entities rather than learning generic syntax and grammar patterns; this may lead to better performance on downstream clinical and biomedical classification tasks which are often knowledge-oriented. Additionally, [8], [9] adapt BERT for long texts without requiring hierarchical splitting methods, which may allow the model to learn useful patterns over longer distances. Lastly, recent work [44] shows that a significant weakness of BioBERT and BlueBERT is that they utilize the original WordPiece vocabulary from BERT, generated from Wikipedia and BooksCorpus; building the WordPiece vocabulary directly on the domain of interest prevents important keywords from being split into multiple subtokens and leads to higher accuracy in downstream tasks. Unfortunately, these approaches have yet to be pretrained on clinical corpora and then released for public use, and thus we leave further evaluation of these methods for future work. The code used for the experiments in our paper will be made available online after peer review.
Acknowledgment
This work has been supported in part by the Joint Design of Advanced Computing Solutions for Cancer (JDACS4C) program established by the U.S. Department of Energy (DOE) and the National Cancer Institute (NCI) of the National Institutes of Health. This work was performed under the auspices of the U.S. Department of Energy by Argonne National Laboratory under Contract DE-AC02-06-CH11357, Lawrence Livermore National Laboratory under Contract DEAC52-07NA27344, Los Alamos National Laboratory under Contract DE-AC5206NA25396, and Oak Ridge National Laboratory under Contract DE-AC05-00OR22725 This research used resources of the Oak Ridge Leadership Computing Facility at the Oak Ridge National Laboratory, which is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC05-00OR22725. This research was supported by the Exascale Computing Project (17-SC-20-SC), a collaborative effort of the U.S. Department of Energy Office of Science and the National Nuclear Security Administration.
Appendix
A. Bootstrapping Confidence Intervals
For each model/task, save the model’s predictions on the test set (hereon referred to as the original predictions)
Randomly select samples from the test set along with their predicted labels (with replacement) to create a new set of samples and predicted labels of the same size as the original test set (hereon referred to as bootstrapped set)
For cancer pathology reports, calculate accuracy and macro F1 score on bootstrapped set; for MIMIC-III, calculate precision, recall, and F1 score on bootstrapped set
Repeat steps (2) and (3) 1000 times, saving the scores each time
Calculate the 95% confidence interval for each metric by finding the 2.5 and 97.5 percentile entry for that metric within the 1000 runs (since precision, recall, and F1 score are not normally distributed)
B. Data Preprocessing
Replace hex and unicode characters with their string equivalents, removing any corrupted codes
For pathology reports, remove identifier segments (registry ID, patient ID, document ID, etc) and XML tags
For MIMIC-III, replace all deidentifier tokens (e.g., [**NAME**]) with the string “deindentified”
Lowercase
Replace all instances of decimal values with the string “floattoken”
Replace all integers higher than 100 with the string “largeinttoken”
Replace all nonalphanerics other than { . ? ! , : ; ( ) % / − + = } with a space
If the same non-alphanumeric character repeats consecutively, replace it with a single copy of that character
Add a space before and after every non-alphanumeric character
C. Attention Visualizations
Fig. A1:

Attention weights and predictions on an example document from the MIMIC-III dataset for the diagnostic category task. In this figure, we multiply the attention weights through both hierarchies of the HiSAN and show exactly which words the HiSAN focuses on in each document (first 510 WordPiece tokens only). For this visualization, we sum the attention weights across all attention heads.
Fig. A2:

Attention weights and predictions on an example document from the MIMIC-III dataset for the diagnostic category task. In the top, using our fine-tuned BlueBERT model (first 510 WordPiece tokens only), we visualize the attention weights from the very final layer that are associated with the [CLS] token used for classification; these weights represent the most important subword tokens after they have already incorporated contextual information from other subword tokens based off the 12 self-attention layers of the main BERT model. In the bottom, we start from the attention weights from the very final layer that are associated with the [CLS] token and multiply these attention weights through all 12 self-attention layers of the BERT model; these weights represent the most important subword tokens accounting for all the inter-word relationships captured during pretraining and fine-tuning. For this visualization, we sum the attention weights across all attention heads.
Footnotes
References
- [1].Wu S, Roberts K, Datta S, Du J, Ji Z, Si Y, Soni S, Wang Q, Wei Q, Xiang Y, Zhao B, and Xu H, “Deep learning in clinical natural language processing: a methodical review.” Journal of the American Medical Informatics Association, vol. 27, no. 3, pp. 457–470, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Devlin J, Chang M-W, Lee K, and Toutanova K, “Bert: Pre-training of deep bidirectional transformers for language understanding,” in NAACLHLT 2019: Annual Conference of the North American Chapter of the Association for Computational Linguistics, 2019, pp. 4171–4186. [Google Scholar]
- [3].Shoeybi M, Patwary M, Puri R, LeGresley P, Casper J, and Catanzaro B, “Megatron-lm: Training multi-billion parameter language models using model parallelism,” arXiv preprint arXiv:1909.08053, 2019. [Google Scholar]
- [4].Collobert R and Weston J, “A unified architecture for natural language processing: deep neural networks with multitask learning,” in Proceedings of the 25th international conference on Machine learning, 2008, pp. 160–167. [Google Scholar]
- [5].Johnson AE, Pollard TJ, Shen L, Lehman L. wei H., Feng M, Ghassemi M, Moody B, Szolovits P, Celi LA, and Mark RG, “Mimic-iii, a freely accessible critical care database,” Scientific Reports, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, and Kang J, “Biobert: a pre-trained biomedical language representation model for biomedical text mining.” Bioinformatics, vol. 36, no. 4, pp. 1234–1240, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Peng Y, Yan S, and Lu Z, “Transfer learning in biomedical natural language processing: An evaluation of bert and elmo on ten benchmarking datasets.” in Proceedings of the 18th BioNLP Workshop and Shared Task, 2019, pp. 58–65. [Google Scholar]
- [8].Kitaev N, Kaiser L, and Levskaya A, “Reformer: The efficient transformer,” in ICLR 2020 : Eighth International Conference on Learning Representations, 2020. [Google Scholar]
- [9].Beltagy I, Peters ME, and Cohan A, “Longformer: The long-document transformer,” arXiv preprint arXiv:2004.05150, 2020. [Google Scholar]
- [10].Qiu JX, Yoon H-J, Fearn PA, and Tourassi GD, “Deep learning for automated extraction of primary sites from cancer pathology reports,” IEEE Journal of Biomedical and Health Informatics, vol. 22, no. 1, pp. 244–251, 2018. [DOI] [PubMed] [Google Scholar]
- [11].Gao S, Qiu JX, Alawad M, Hinkle JD, Schaefferkoetter N, Yoon H-J, Christian B, Fearn PA, Penberthy L, Wu X-C, Coyle L, Tourassi G, and Ramanathan A, “Classifying cancer pathology reports with hierarchical self-attention networks.” Artificial Intelligence in Medicine, vol. 101, p. 101726, 2019. [DOI] [PubMed] [Google Scholar]
- [12].Talmor A and Berant J, “Multiqa: An empirical investigation of generalization and transfer in reading comprehension,” in ACL 2019 : The 57th Annual Meeting of the Association for Computational Linguistics, 2019, pp. 4911–4921. [Google Scholar]
- [13].Alberti C, Lee K, and Collins M, “A bert baseline for the natural questions,” arXiv preprint arXiv:1901.08634, 2019. [Google Scholar]
- [14].Petroni F, Rocktschel T, Riedel S, Lewis P, Bakhtin A, Wu Y, and Miller A, “Language models as knowledge bases,” in 2019 Conference on Empirical Methods in Natural Language Processing, 2019, pp. 2463–2473. [Google Scholar]
- [15].Soares LB, FitzGerald N, Ling J, and Kwiatkowski T, “Matching the blanks: Distributional similarity for relation learning,” in ACL 2019: The 57th Annual Meeting of the Association for Computational Linguistics, 2019, pp. 2895–2905. [Google Scholar]
- [16].Liu Y and Lapata M, “Text summarization with pretrained encoders,” in 2019 Conference on Empirical Methods in Natural Language Processing, 2019, pp. 3728–3738. [Google Scholar]
- [17].Zhang H, Xu J, and Wang J, “Pretraining-based natural language generation for text summarization.” arXiv preprint arXiv:1902.09243, 2019. [Google Scholar]
- [18].Sun C, Qiu X, Xu Y, and Huang X, “How to fine-tune bert for text classification?” China National Conference on Chinese Computational Linguistics, pp. 194–206, 2019. [Google Scholar]
- [19].Adhikari A, Ram A, Tang R, and Lin J, “Docbert: Bert for document classification,” arXiv preprint arXiv:1904.08398, 2019. [Google Scholar]
- [20].Reimers N, Schiller B, Beck T, Daxenberger J, Stab C, and Gurevych I, “Classification and clustering of arguments with contextualized word embeddings,” in ACL 2019 : The 57th Annual Meeting of the Association for Computational Linguistics, 2019, pp. 567–578. [Google Scholar]
- [21].Ostendorff M, Bourgonje P, Berger M, Schneider JM, Rehm G, and Gipp B, “Enriching bert with knowledge graph embeddings for document classification.” KONVENS, 2019. [Google Scholar]
- [22].Alsentzer E, Murphy JR, Boag W, Weng W-H, Jin D, Naumann T, and McDermott MBA, “Publicly available clinical bert embeddings.” arXiv preprint arXiv:1904.03323, 2019. [Google Scholar]
- [23].Martina S, “Classification of cancer pathology reports with deep learning methods,” Ph.D. dissertation, 2020. [DOI] [PubMed] [Google Scholar]
- [24].Sheikhalishahi S, Miotto R, Dudley JT, Lavelli A, Rinaldi F, and Osmani V, “Natural language processing of clinical notes on chronic diseases: Systematic review.” JMIR medical informatics, vol. 7, no. 2, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Solares JRA, Raimondi FED, Zhu Y, Rahimian F, Canoy D, Tran J, Gomes ACP, Payberah AH, Zottoli M, Nazarzadeh M, Conrad N, Rahimi K, and Salimi-Khorshidi G, “Deep learning for electronic health records: A comparative review of multiple deep neural architectures,” Journal of Biomedical Informatics, vol. 101, p. 103337, 2020. [DOI] [PubMed] [Google Scholar]
- [26].Lee J, Song H-J, Yoon E, Park S-B, Park S-H, Seo J-W, Park P, and Choi J, “Automated extraction of biomarker information from pathology reports,” BMC medical informatics and decision making, vol. 18, no. 1, p. 29, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Xie F, Lee J, Munoz-Plaza CE, Hahn EE, and Chen W, “Application of text information extraction system for real-time cancer case identification in an integrated healthcare organization,” Journal of pathology informatics, vol. 8, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yala A, Barzilay R, Salama L, Griffin M, Sollender G, Bardia A, Lehman C, Buckley JM, Coopey SB, Polubriaginof F, et al. , “Using machine learning to parse breast pathology reports,” Breast cancer research and treatment, vol. 161, no. 2, pp. 203–211, 2017. [DOI] [PubMed] [Google Scholar]
- [29].Yim W.-w., Denman T, Kwan SW, and Yetisgen M, “Tumor information extraction in radiology reports for hepatocellular carcinoma patients,” AMIA Summits on Translational Science Proceedings, vol. 2016, p. 455, 2016. [PMC free article] [PubMed] [Google Scholar]
- [30].Gehrmann S, Dernoncourt F, Li Y, Carlson ET, Wu JT, Welt J, Foote J, Moseley ET, Grant DW, Tyler PD, and Celi LA, “Comparing deep learning and concept extraction based methods for patient phenotyping from clinical narratives.” PLOS ONE, vol. 13, no. 2, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mullenbach J, Wiegreffe S, Duke J, Sun J, and Eisenstein J, “Explainable prediction of medical codes from clinical text,” in NAACLHLT 2018: 16th Annual Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, vol. 1, 2018, pp. 1101–1111. [Google Scholar]
- [32].Baumel T, Nassour-Kassis J, Cohen R, Elhadad M, and Elhadad N, “Multi-label classification of patient notes a case study on icd code assignment,” in AAAI Workshops, 2017, pp. 409–416. [Google Scholar]
- [33].Gao S, Young MT, Qiu JX, Yoon H-J, Christian JB, Fearn PA, Tourassi GD, and Ramanthan A, “Hierarchical attention networks for information extraction from cancer pathology reports,” J Am Med Inform Assoc, vol. 25, no. 3, pp. 321–330, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Qiu X, Sun T, Xu Y, Shao Y, Dai N, and Huang X, “Pre-trained models for natural language processing: A survey,” arXiv preprint arXiv:2003.08271, 2020. [Google Scholar]
- [35].Liu Q, Kusner MJ, and Blunsom P, “A survey on contextual embeddings,” arXiv preprint arXiv:2003.07278, 2020. [Google Scholar]
- [36].Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser L, and Polosukhin I, “Attention is all you need,” in Proceedings of the 31st International Conference on Neural Information Processing Systems, 2017, pp. 5998–6008. [Google Scholar]
- [37].Kim Y, “Convolutional neural networks for sentence classification,” in Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), 2014, pp. 1746–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Young T, Hazarika D, Poria S, and Cambria E, “Recent trends in deep learning based natural language processing,” arXiv preprint arXiv:1708.02709, 2017. [Google Scholar]
- [39].Xiao C, Choi E, and Sun J, “Opportunities and challenges in developing deep learning models using electronic health records data: a systematic review,” Journal of the American Medical Informatics Association, vol. 25, no. 10, pp. 1419–1428, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hughes M, Li I, Kotoulas S, and Suzumura T, “Medical text classification using convolutional neural networks,” Stud Health Technol Inform, vol. 235, pp. 246–250, 2017. [PubMed] [Google Scholar]
- [41].Alawad M, Gao S, Qiu JX, Yoon HJ, Blair Christian J, Penberthy L, Mumphrey B, Wu X-C, Coyle L, and Tourassi G, “Automatic extraction of cancer registry reportable information from free-text pathology reports using multitask convolutional neural networks,” Journal of the American Medical Informatics Association, vol. 27, no. 1, pp. 89–98, 11 2019. [Online]. Available: 10.1093/jamia/ocz153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zhang Y, Chen Q, Yang Z, Lin H, and Lu Z, “Biowordvec, improving biomedical word embeddings with subword information and mesh.” Scientific Data, vol. 6, no. 1, p. 52, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Xiong W, Du J, Wang WY, and Stoyanov V, “Pretrained encyclopedia: Weakly supervised knowledge-pretrained language model,” in ICLR 2020 : Eighth International Conference on Learning Representations, 2020. [Google Scholar]
- [44].Gu Y, Tinn R, Cheng H, Lucas M, Usuyama N, Liu X, Naumann T, Gao J, and Poon H, “Domain-specific language model pretraining for biomedical natural language processing.” arXiv preprint arXiv:2007.15779, 2020. [Google Scholar]


