FIGURE 3.
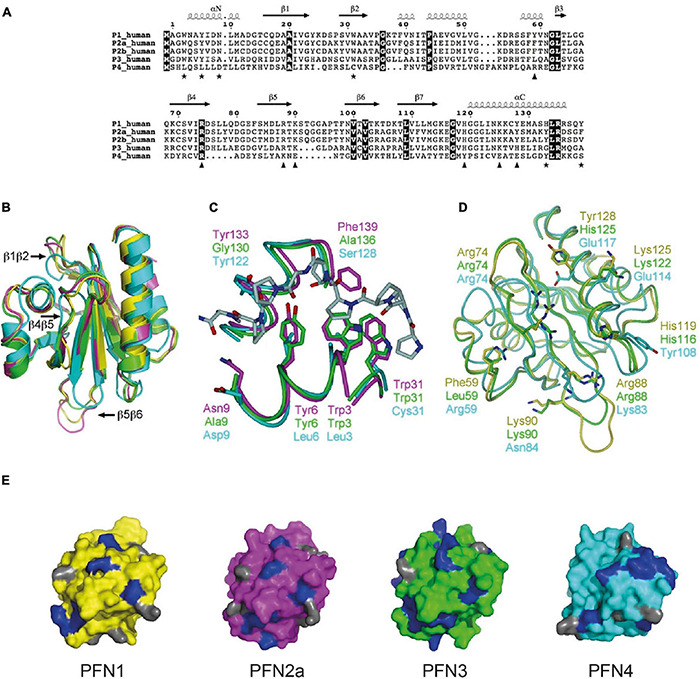
Structural features reflect the different biochemical properties of the mammalian profilin isoforms. (A) Sequence alignment between human profilins. Fully conserved residues are on dark background. Key residues of PLP binding are indicated by asterisks, those involved in binding actin with triangles. Secondary structures are derived from bovine PFN1 crystal structure. For compatibility with most publications, the first methionine is not considered in sequence numbering. (B) Superposition of bovine PFN1 (yellow), mouse PFN2a (magenta), human PFN3 (green), and human PFN4 (cyan). PFN1 structure is from the profilactin complex (PDB core 1HLU) and PFN2a from the complex with a VASP peptide (PDB code 2V8C). Loops variable in length in PFN3 and/or PFN4 are marked. (C) Comparison of the PLP-binding sites of PFN2a, PFN3, and PFN4. Shown is also the PFN2a-VASP complex, coloring as in 3B. Only half of the binding site is conserved in PFN3, no conservation is seen in PFN4. Key residues for peptide binding are indicated. (D) Actin-binding sites of PFN1, PFN3, and PFN4, coloring as in 3B. For clarity, actin is not shown; view is from the direction of actin onto the actin-binding surface on profilin, side chains of key profilin residues are shown. (E) Comparison of PtdIns(4,5)P2 binding surfaces of PFN1, 2a, 3, and 4 (left to right, respectively). Profilins are colored as in 3A, and all arginine residues, crucial for PtdIns(4,5)P2 binding, are highlighted in blue; lysine residues are shown in gray. This figure is used in this review with permission from Behnen et al. (2009).
