FIGURE 6.
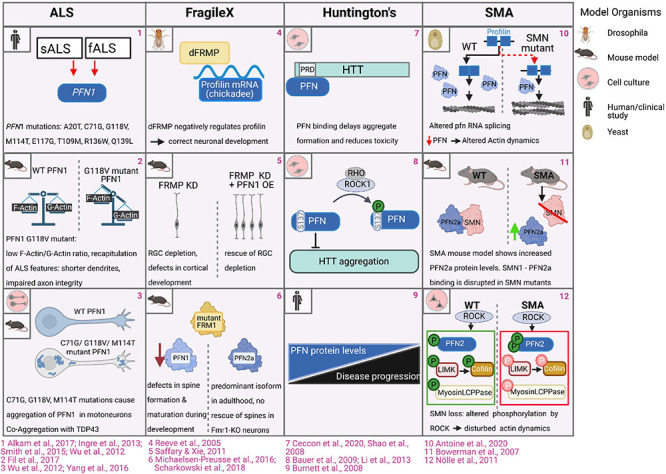
Profilins in CNS pathologies. 1. Exome sequencing studies revealed 8 mutations in the Pfn1 gene in familial and sporadic ALS patients (Ingre et al., 2013; Wu et al., 2012; Chen et al., 2013; Smith et al., 2015; Alkam et al., 2017). 2. Motoneurons expressing ALS mutant PFN1 G188V exhibit abnormally low ratios of F-/G-actin. Motor neurons of ALS-PFN1-mutant expressing mouse model exhibited cytoskeletal disruptions and impaired axon integrity followed by Wallerian degeneration (Yang et al., 2016). 3. In cell culture and mice, wildtype PFN1 is diffusely distributed across the cytoplasm of motor neurons and N2A cells, while PFN1 C71G, PFN1 M114T and PFN1 G118V PFN mutations form protein aggregates and inclusions. PFN1 co-aggregates with TDP43 (Wu et al., 2012; Yang et al., 2016). 4. FRMP directly binds the mRNA of Drosophila profilin (encoded by chickadee), which is required for correct neuronal development in Drosophila (Reeve et al., 2005). 5. FRMP knockdown in mice causes depletion of radial glia cells (RGC) and defects in cortical development, while overexpression of PFN1 in these settings restores the radial glial dependent developmental defects (Saffary and Xie, 2011). 6. Levels of PFN1, but not PFN2a, are reduced in FRM1 mutant mice. Depletion of PFN1 causes defective dendritic spine formation and maturation during development analogous to defects in Fmr1-KO mice. Overexpressed PFN1, but not PFN2a, can rescue dendritic spine defects in Fmr1-KO neurons (Michaelsen et al., 2010; Scharkowski et al., 2018). 7. Binding of PFN to the proline-rich domain of huntingtin reduces huntingtin aggregation and toxicity (Shao et al., 2008; Ceccon et al., 2020). 8. Rho kinase-dependent phosphorylation of PFN1 at serine 137/8 enhances huntingtin aggregation. Pharmacological inhibition of ROCK inhibits huntingtin aggregation and toxicity (Bauer et al., 2009; Li et al., 2013). 9. In humans, reduced profilin protein levels correlate with disease progression of Huntington disease in patients (Burnett et al., 2008). 10. smn-deficient fission yeast displays splicing defects in the profilin gene, resulting in altered actin network homeostasis, perturbed cytokinesis and endocytosis (Antoine et al., 2020). 11. Loss of SMN in SMA settings is associated with the upregulation of PFN2a protein levels (Bowerman et al., 2007). 12. Levels of phosphorylated PFN2a are increased in SMA settings while ROCK-dependent phosphorylation of myosin light chain protein phosphatase (Myosin LCPPase) and the cofilin-regulating LIM-kinase, are reduced, causing disturbed actin dynamics (Nölle et al., 2011).
