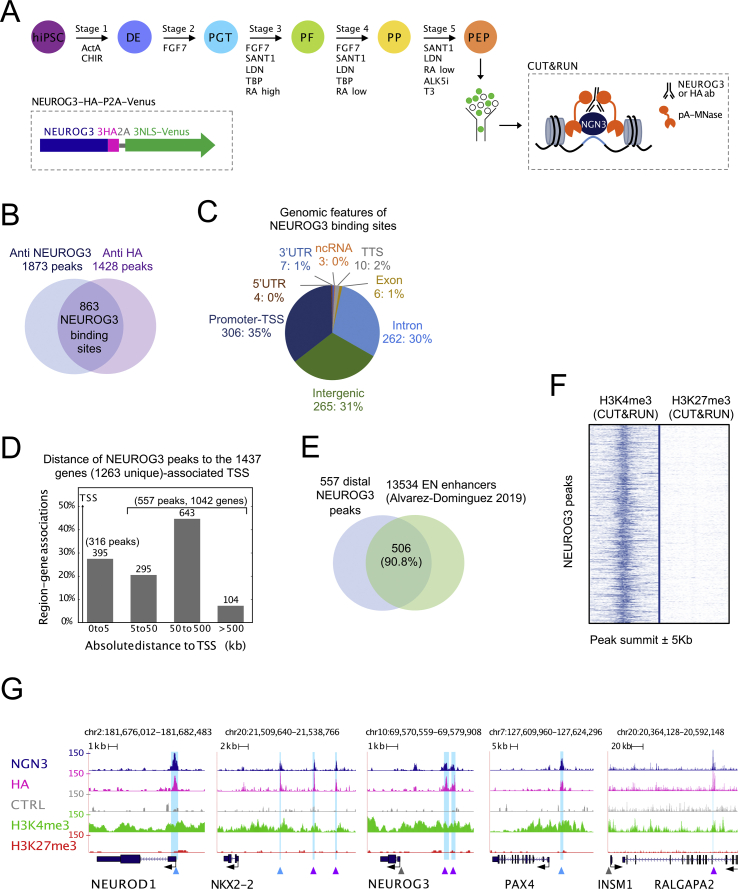
Figure 1.
Characterization of the genome-wide binding sites of NEUROG3 in human hiPSC-derived pancreatic endocrine progenitors. (A) Overview of the study: a 5-stage protocol was used to differentiate hiPSC to pancreatic endocrine progenitors using the sequential supplementation of factors indicated. At day 13, Venus+ cells were sorted and used in a CUT&RUN experiment. Inset: schematic representation of the NEUROG3-3HA-P2A-3NLS-Venus allele. (B) Venn diagram showing the number and overlap of peaks identified by CUT&RUN with an anti-NEUROG3 or an anti-HA antibody. (C) Genomic distribution (number and % of peaks) of the 863-high confidence NEUROG3 binding sites. (D) Distance of NEUROG3 peaks to their gene(s)-associated TSS. (E) Overlap between NEUROG3 distal binding sites (>5 kb from TSS) and enhancers regions of hiPSC-derived endocrine progenitors (EN), as defined by Alvarez-Dominguez et al. [30], P = 4.9e-324. (F) Normalized read density surrounding NEUROG3 peak summit ±5 Kb for H3K4me3 and H3K27me3 CUT&RUN datasets. (G) Genome browser tracks showing NEUROG3, HA, H3K4me3, H3K27me3, and the CTRL (Donkey anti-Sheep antibody) CUT&RUN data at the NEUROD1, NKX2-2, NEUROG3, PAX4, and INSM1/RALGAPA2 loci. Identified NEUROG3 peaks are highlighted in light blue. Peaks matching previously reported NEUROG3 binding sites are indicated by blue arrowheads, newly discovered peaks by purple arrowheads, and reported sites not confirmed here by gray arrowheads [12,13,[15], [16], [17]].