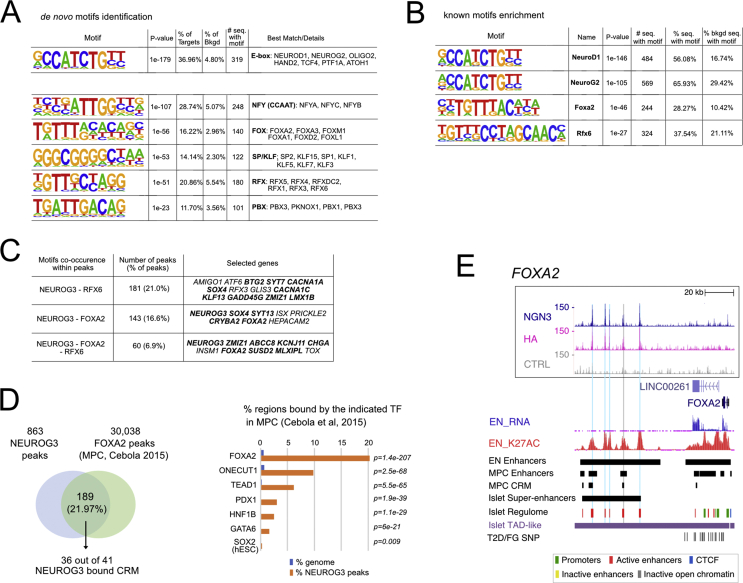
Figure 2.
TF motifs discovery in NEUROG3 binding sites. (A). De novo motif discovery ranked by P-value, reflecting motif enrichment within peak summit ±100 bp. Number (#) and % of target and background sequences harboring each motif is indicated for the 6 most significant motifs identified. Known best matching transcription factors were associated if their HOMER score was >0.85. (B) Selection of known co-occurring motifs ranked by P-value within the entire peak sequences. The full list of the 50 most significantly enriched motifs is shown in Suppl Figure 4. (C) Co-occurrence of NEUROG3 de novo–identified motifs with motifs for RFX6 and/or FOXA2 on the entire peak sequences. Some selected GREAT assigned genes are indicated, in bold when identified as targets for Rfx6 in a mouse beta cell line [45]. (D) Regions bound by NEUROG3 at PEP stage that are bound by FOXA2 and indicated TFs at the MPC stage. NEUROG3 cistrome was intersected with that of FOXA2, ONECUT1, TEAD1, GATA6, PDX1, and HNF1b from in vitro–derived MPC and, as a control, with that of SOX2 from hESC cells, taken from Ref. [31]. P-value of enrichment at NEUROG3 binding sites relative to genomic distribution is calculated by CEAS. In the left panel, the number of NEUROG3-bound and NEUROG3/FOXA2-bound CRMs is indicated. (E) Genome browser tracks showing NEUROG3, HA, and the CTRL CUT&RUN data at the FOXA2 loci. Coordinates are from hg19. The positions of NEUROG3 binding sites are highlighted in light blue (or in gray, when identified in a single dataset). Data of RNA-seq (EN_RNA), H3K27ac ChIP-seq (EN_K27AC), and the position of enhancers (EN Enhancers) from hESC-differentiated to endocrine progenitors were taken from Ref. [30] and http://meltonlab.rc.fas.harvard.edu/data/UCSC/SCbetaCellDiff_ATAC_H3K4me1_H3K27ac_WGBS_tracks.txt. Position of hESC-derived multipotent progenitor cells (MPC) enhancers and cis-regulatory modules (CRM) are taken from Ref. [31]. Data from adult islets (Super-enhancers, Islet regulome, T2D/FG SNP and TAD-like regions) are taken from Ref. [32], isletregulome.org and http://epigenomegateway.wustl.edu/.