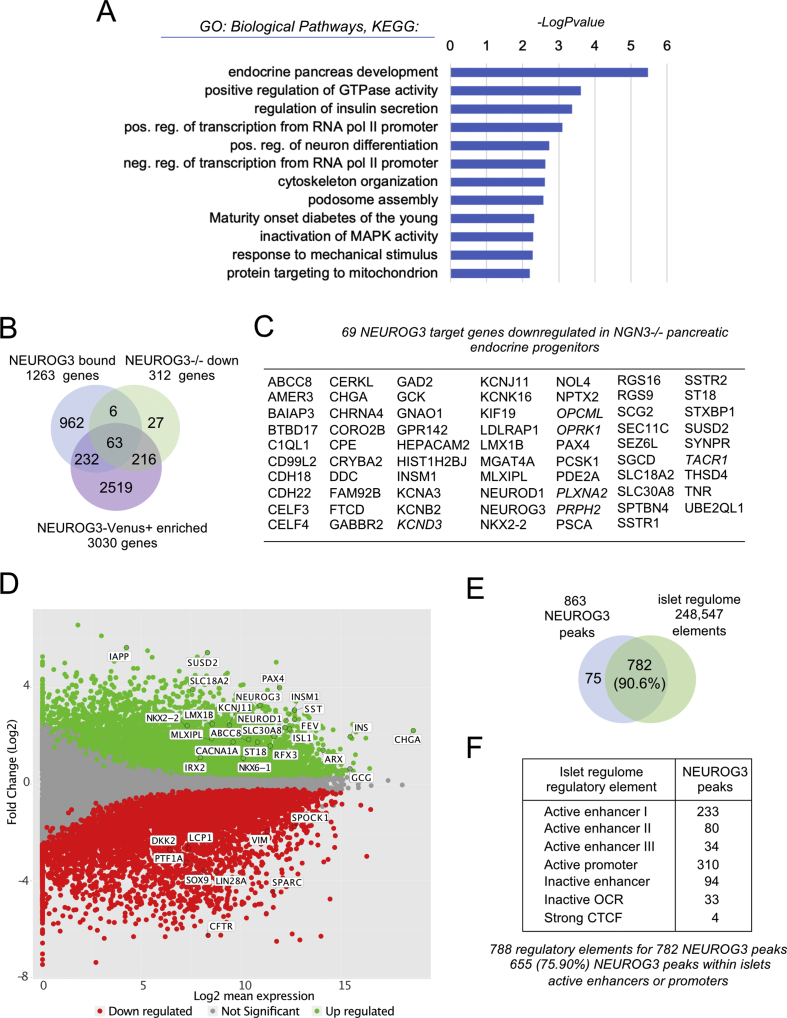
Figure 3.
Integration of NEUROG3 occupancy and islet expression of target candidates. (A) Gene ontology analysis (biological process, KEGG pathways) showing selected significantly enriched terms (Log10[P-value] ≥ 2) related to the 1263 NEUROG3 bound genes. (B) Venn diagram illustrating the overlap of NEUROG3 bound genes and the 312 downregulated genes in NEUROG3−/− hESC line differentiated to PEP stage (NEUROG3−/− down; [25]) and the 3030 genes enriched in NEUROG3-HA-P2A-Venus+ compared to Venus− cells differentiated to PEP stage (NEUROG3-Venus+ enriched). (C) List of the 69 NEUROG3 target genes downregulated in NEUROG3−/− PEP. In italics, the 6 bound genes downregulated in NEUROG3−/− PEP but not enriched in NEUROG3-Venus+ PEP. (D) MA-plot representing the estimated Log2 fold change as a function of the Log2 mean expression of genes enriched in NEUROG3-Venus+ (green) or NEUROG3-Venus− (red) cells. (E–F) NEUROG3 binding sites matching with one or more islets regulatory element(s) (islet regulome; [32]). OCR, open chromatin regions, CTCF, CTCF binding sites.