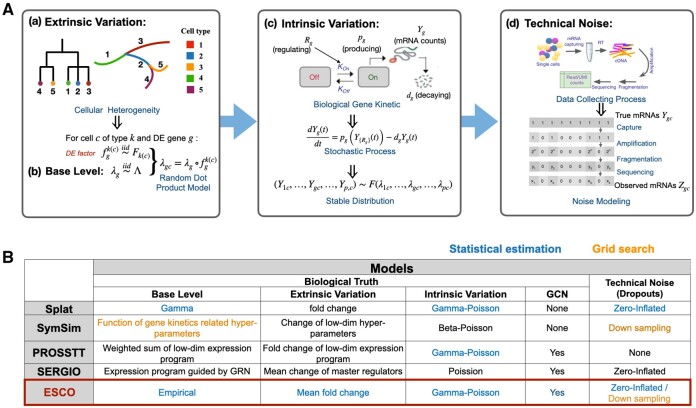
Fig. 1.
Summary of simulators for scRNA-seq data. (A) The general modeling flowchart of commonly used simulators. Simulators often start with (a) extrinsic variation that arose from cell heterogeneity in the biological sense, and import this model to (b) the base expression mean generated for each gene, to formalize the heterogeneous expression means for a gene in a cell of a particular cell type. Then, those means are used to generate the expression level, i.e. mRNA counts, by modeling the (c) intrinsic variation, i.e. the stochasticity of gene expression in a cell with a defined base rate of expression. This process is often modeled by the gene kinetic model in biochemistry, which could be stated as a stochastic process in statistical terms. The stable distribution of this stochastic process can usually be approximated by distributions like negative binomial/Poisson/beta Poisson. Finally, some simulators allow the generation of technical noise (d) separately, by adding noise, step by step, to the true counts, to mimic the data collection process [the cartoon display is from Zhang et al. (2019a)]. Usually, this stepwise process is approximated by the zero-inflation model, where the true counts are set to zero with probability related to expression level. (B) Summary of the current state of simulators following the general modeling flowchart described above, with blue and orange text color indicating whether they use statistical estimation or grid search when fitting the simulator to a real dataset. The objective of ESCO is to create an ensemble of the best features among current simulators in each step, while allowing easily imposing co-expression structure among genes via a copula